AI-Powered Breakthroughs in Antibody Optimization
Artificial intelligence (AI) is a game changer for developing better antibody treatments. AI can analyze and improve antibodies, replacing the traditional time-consuming and expensive methods and making lead optimization faster and more effective. In this blog, we highlight three main ways AI can be used to enhance the lead optimization efforts, depending on the type of data available: the antibody sequence, the sequence and structure of antibody, or comprehensive data including the sequence and structure of both antibody and its’ target. Through real-world examples and specific AI tools, we demonstrate how AI can not only suggest improvements to existing antibodies but also generate new ones that could work even better.
Table of Contents:
- AI-Powered Antibody Optimization: The Art of Precision in Lead Optimization
- Tailored Computational Strategies for Unmatched Potential
- AI-driven Antibody Optimization Based On Sequence
- AI-Driven Antibody Optimization Based on Sequence and Structure
- AI-Driven Antibody Optimization Based on Binder-Target Structural Context
- The Intersection of AI and Human Expertise in Antibody Optimization
AI-Powered Antibody Optimization: The Art of Precision in Lead Optimization
In drug discovery, lead optimization is a crucial phase where potential therapies, especially monoclonal antibodies (mAbs), undergo meticulous refinement. The difference between a good and an exceptional therapeutic often relies on incredibly subtle molecular modifications.
Traditional lead optimization practices, especially the ones focusing on affinity maturation, involve subjecting candidate antibodies to a laborious and costly in vitro affinity maturation process. AI-based solutions streamline and enhance the antibody discovery process, reducing the time and increasing the success rate of lead optimization efforts.
Tailored Computational Strategies for Unmatched Potential
The power of AI-based methods lies in their ability to utilize any type of available data. Depending on the input type, three distinct computational strategies can be applied:
- Utilizing the antibody sequence alone.
- Harnessing both the antibody sequence and its structure.
- Leveraging the sequence and structural data of the antibody and its target.
Each data type unlocks specific AI capabilities, providing unique insights and paving the way toward novel therapeutic discoveries. At Ardigen, we develop cutting-edge AI-driven in-silico methodologies to empower data-driven drug discovery. Follow along to discover how we redefine the boundaries of therapeutic innovation.

AI-driven Antibody Optimization Based On Sequence
In cases when only the antibody sequence is available, AI algorithms based on large language models (LLMs) adapted for analyzing protein sequences present a novel avenue for antibody design. A recent study by Hie, B.L. et al., published in Nature Biotechnology in 2023 demonstrated that such models can suggest beneficial mutations for improving antibodies in the absence of detailed structural information or knowledge of the target antigen.
The authors applied a language-model-driven process to the affinity maturation of seven antibodies targeting, among others, SARS-CoV-1, SARS-CoV-2, Ebola, and influenza viruses. By examining no more than 20 variants per antibody over just two iterations of AI-aided computational design, they were able to enhance the binding affinity of four clinically significant, well-developed antibodies by as much as seven-fold. They also achieved up to a 160-fold increase in affinity for three antibodies that were less mature with the best results for the one targeting the Ebola virus. Notably, several of the AI-optimized antibodies exhibited improved thermostability and showed promising activity in neutralizing viruses, specifically the Ebola and SARS-CoV-2 pseudoviruses. These remarkable results illustrate the potential of AI to streamline the antibody development process using protein sequence information alone.
The study also suggests that the underlying models that enhance antibody affinity could be broadly applied to other protein engineering efforts. This general approach, from sequence analysis to optimized leads, is outlined below.

Several sequence-based AI models have been developed specifically for antibody optimization, each featuring unique strengths. For example, the ESM2 and ProtT5 models offer a broad-spectrum analysis based on general protein evolution, while AbLang specializes in more specific details of antibody sequences. Complementing these is Ardigen’s Prism, which is a curated collection of protein LLMs that brings together the collective power of multiple models to support and enhance precision protein engineering.
AI-Driven Antibody Optimization Based on Sequence and Structure
When both the antibody sequence and structure are available, scientists can capitalize on the value of structural insight. This approach employs various AI models that take the 3D protein structure as input and predict sequences with a high probability of folding into the given shape. Given the initial 3D structure of an antibody, this method focuses on the regions known as the Complementarity-Determining Regions (CDRs), which are essential for antigen binding. The original CDR sequences are then ‘masked’, allowing the models to generate a range of novel sequences anticipated to fold in a manner similar to the original antibody. Below is a schematic of this approach:
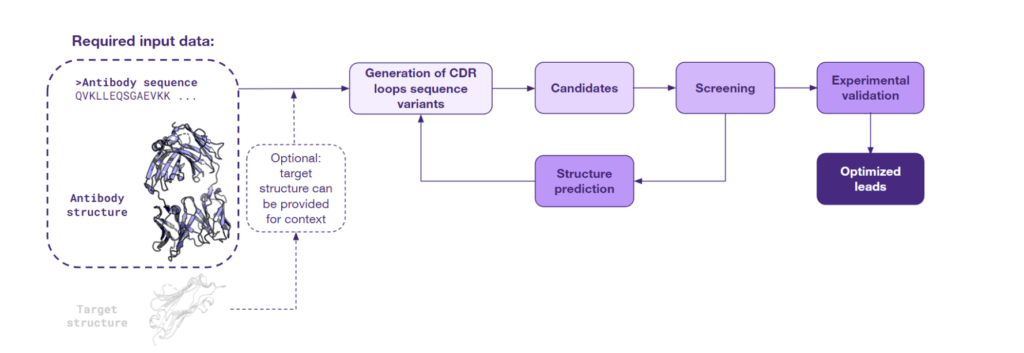
This process often enables the regeneration of the original sequence and creation of diverse novel variants. The novel variants can display similar or even improved affinity to the original target and potential cross-reactivity with similar targets, thereby expanding the therapeutic potential of the antibody.
The efficacy of the inverse folding approach has been demonstrated by Frederic A. Dreyer et al., in the manuscript published in 2023 on arXiv. The study develops a deep learning inverse folding model specifically adapted for antibody sequence design. The AbMPNN model specifically trained to take into account the target structure as context has set new benchmarks for antibody designability, particularly for the hypervariable CDR-H3 loop. This model belongs to a suite of computational tools designed for sequence regeneration from a given structure, including others like ProteinMPNN.
The results of the study underscore the high impact of integrating sequence and structural data for lead optimization. Intriguingly, the inverse folding method can be utilized even when an antibody’s experimental structure remains unknown. Thanks to recent advances in antibody structure prediction, a modeled structure derived from its sequence can serve as a starting point for optimization. Moreover, sequences developed in this way can undergo the sequence diversification process described above, potentially expanding the arsenal of antibodies explored by AI to refine the initial leads.
AI-Driven Antibody Optimization Based on Binder-Target Structural Context
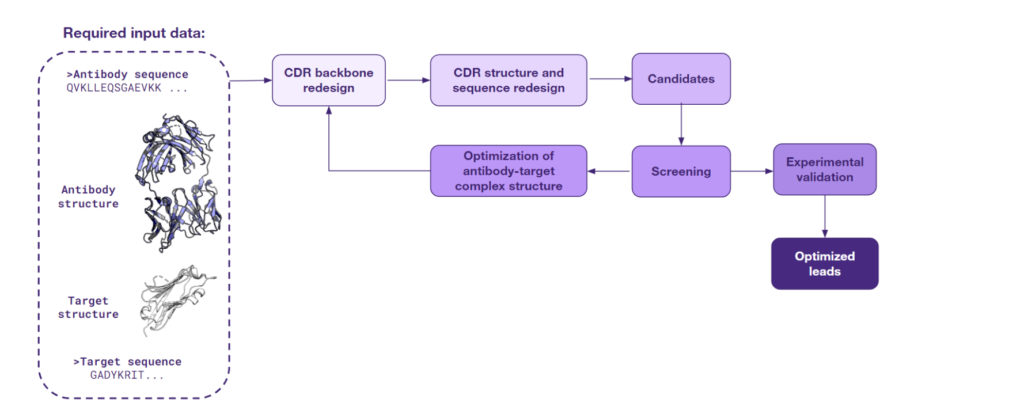
The third approach to lead optimization is at the forefront of innovation. It harnesses the latest AI techniques for de novo generation of protein structures and sequences and is being actively pursued by the scientific community for its groundbreaking potential. This strategy is based on generating protein configurations in specific contexts.
Starting with the structure of an antibody-target complex, this particular type of AI models focus on redesigning the CDRs. These are the regions of the antibody that directly engage with the target and determine the strength and specificity of the antigen binding and, consequently, modulate the immune response. Through the CDR redesigning process, AI is able to design a completely new interface, one that is more likely to exhibit a high specificity for the antigen.
An array of sophisticated models has been developed to support this novel approach. One of them is RFdiffusion, which was showcased by Charlotte Deane at the PEGS 2023 conference in Lisbon. What is more, a recent study from Baker’s group has shown that the RFdiffusion model, fine-tuned for specificity, can create new antibody variable heavy chains that bind precisely to designated epitopes. Other models such as Chroma, AbDesign & AbDock, and EAGLE also bring unique capabilities to the table. These models specialize in the de novo generation of protein sequences and structures, a task that is energizing the scientific community with its promise and complexity.
The figure below captures the utility of this approach for lead optimization, from initial structure to optimized leads. As AI continues to evolve, such solutions will likely become central to the discovery and refinement of therapeutic antibodies.
The Intersection of AI and Human Expertise in Antibody Optimization
The future of antibody development is defined by the dynamic intersection of computational power and biological innovation. AI methods hold remarkable potential to accelerate and refine the process of lead optimization. However, it’s essential to recognize that these advanced computational tools do not operate in isolation. The true power of AI is realized when it is employed by experts who understand both the science of antibody development and the nuances of machine learning models.
While AI opens new possibilities in antibody lead optimization, it is the human expertise in using these tools that propels successful outcomes. At Ardigen, we leverage our deep understanding of both AI algorithms and biological systems to selectively advance the most promising antibodies. This strategic approach ensures efficient use of resources and timely execution to move us closer to breakthrough therapies. Our team embraces AI innovation in addition to our established scientific acumen to maintain our position as leaders in antibody development.
Works Cited:
- B.L. Hie, et al. (2023). “Efficient evolution of human antibodies from general protein language models,” Nature Biotechnology. Available at: https://www.nature.com/articles/s41587-023-01763-2
- Zeming Lin, et al. (2022). “Language models of protein sequences at the scale of evolution enable accurate structure prediction” BioRxiv. Available at: https://www.biorxiv.org/content/10.1101/2022.07.20.500902v1.full.pdf
- M. Heinzinger, et al. (2024). “Bilingual Language Model for Protein Sequence and Structure” BioRxiv. Available at: https://www.biorxiv.org/content/10.1101/2023.07.23.550085v2
- T.H. Olsen, et al. (2024). “Addressing the antibody germline bias and its effect on language models for improved antibody design” BioRxiv. Available at: https://www.biorxiv.org/content/10.1101/2024.02.02.578678v1
- Ardigen, (2020). “PRISM: A Writing Assistant for the Language of Proteins” Available at: https://ardigen.com/prism-a-writing-assistant-for-the-language-of-proteins-2/
- F.A. Dreyer, et al. (2023). “Inverse folding for antibody sequence design using deep learning” arXiv. Available at: https://arxiv.org/abs/2310.19513
- F.A. Dreyer, et al. (2023). “Inverse folding for antibody sequence design using deep learning” Zenodo. Available at: https://zenodo.org/records/8164693
- J. Dauparas, et al. (2022). “Robust deep learning-based protein sequence design using ProteinMPNN” J. BioRxiv. Available at: https://www.biorxiv.org/content/10.1101/2022.06.03.494563v1
- N. R. Bennett, et al. (2024). “Atomically accurate de novo design of single-domain antibodies” BioRxiv. Available at: https://www.biorxiv.org/content/10.1101/2024.03.14.585103v1
- J.B. Ingraham, et al. (2023). “Illuminating protein space with a programmable generative model” Nature. Available at: https://www.nature.com/articles/s41586-023-06728-8#citeas
- Z. Peng, et al. (2023). “Generative Diffusion Models for Antibody Design, Docking, and Optimization” BioRxiv. Available at: https://www.biorxiv.org/content/10.1101/2023.09.25.559190v1
- T. Cohen and D. Schneidman-Duhovny (2023). “Epitope-specific antibody design using diffusion models on the latent space of ESM embeddings” OpenReview.net. Available at: https://openreview.net/forum?id=Enqxq6TWoZ




