AI for Biologics
Develop Safe, Targeted Therapeutics with a Leading AI CRO
Advance Your Biologics from Concept to Clinical Readiness with AI
Whether you’re generating leads, screening hits, or optimizing candidates, Ardigen’s AI-powered approach is designed to enhance each step. With expertise in amino acid-based therapeutics—like antibodies, TCRs, peptides, and recombinant proteins—we work closely with your data to deliver precise, meaningful outcomes, driving AI-driven biologics development.

Surpass the Limits of Traditional Biologics Development with AI
Accurate Target Characterization for Biologics Efficacy
Ensure efficacy and safety by precisely identifying the epitope, analyzing the target’s biological role, and mitigating potential off-target effects using advanced biologics AI.
Effective lead generation
Increase the probability of finding promising leads by generating a diverse array of biological candidates with high activity and specificity using advanced AI models to expand the screening space, crucial for biologics drug discovery.
Intelligent hit screening
Select the best candidates based on safety (immunogenicity), developability (protein stability, solubility, aggregation), and functionality (affinity, activity).
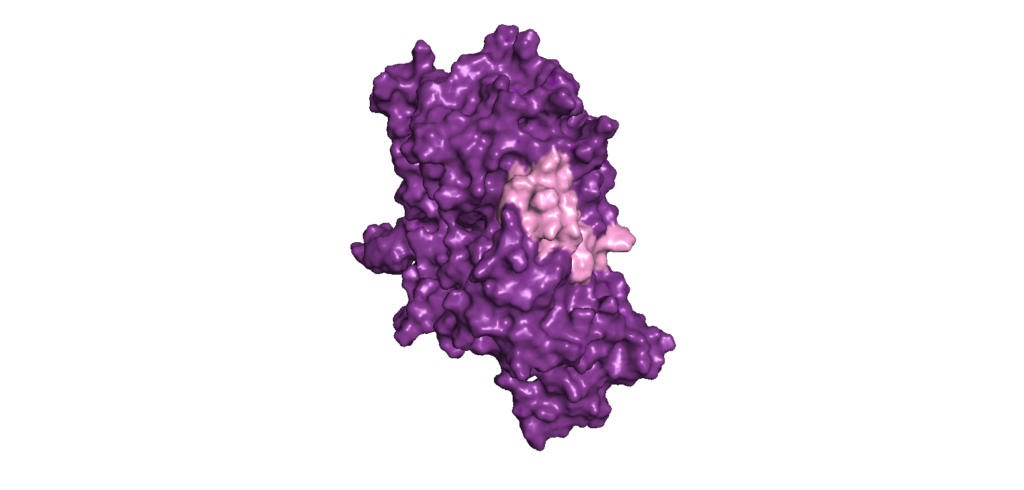
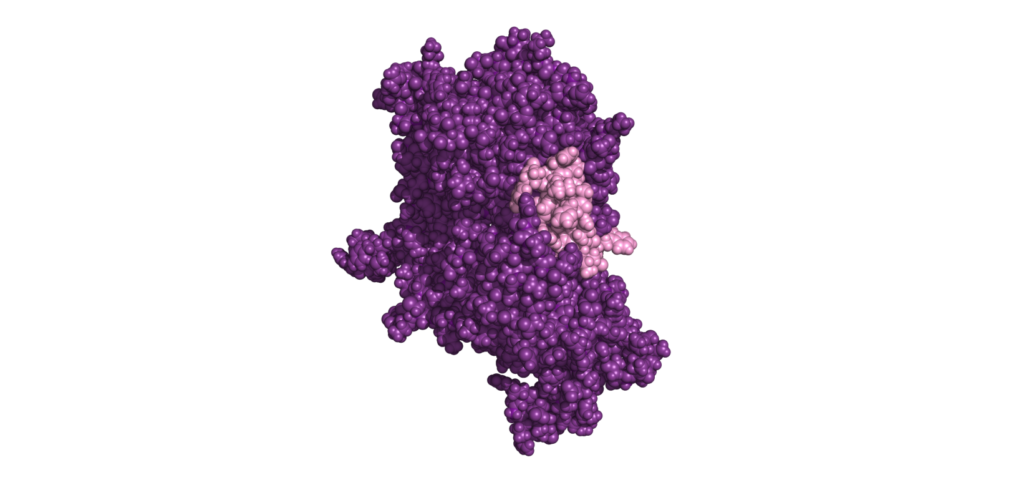
Interaction insights: Binder-Target Analysis for Biologics Safety
Ensure good selective binding and minimize off-target effects by performing Binder-Target Interface Analysis of the molecular interactions, leveraging our expertise in antibody modeling.
AI solutions grounded in biology For Biologics Development
With advanced AI models, state-of-the-art bioinformatics and physics-based tools at your hands, you can develop biologics that are effective, precise and optimized for performance, enabling true AI-driven biologics development.
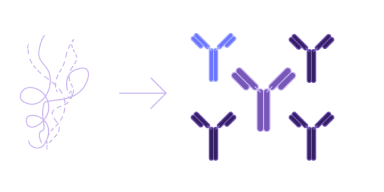
De novo lead generation
Uncover novel and unique candidates for biologics discovery, even for the most challenging targets by using advanced Ai solutions to greatly expand the candidate screening space.
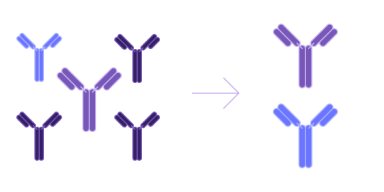
Hit screening
Select the best candidates and ensure optimal efficacy, functionality, and safety by employing pre-trained AI models or developing customized models that leverage your data for enhanced accuracy in biologics drug discovery.
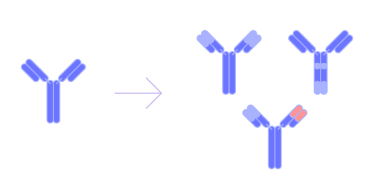
Lead optimization
Develop candidates with improved affinity, activity, specificity, and physicochemical properties by employing high-performance approaches like Large Language Models, inverse folding, and the de novo generation of protein fragments.
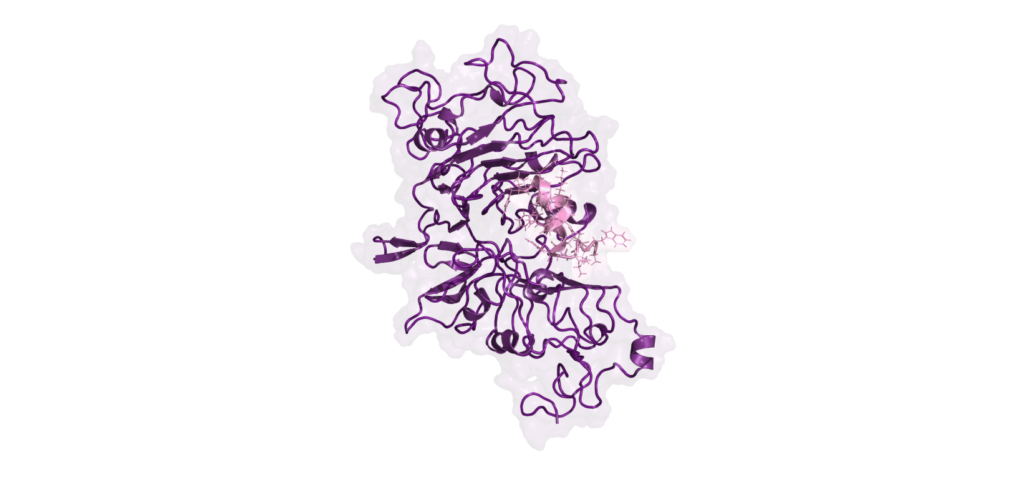
Deep molecular insights
We combine AI for biologics with empirical techniques, such as protein-protein docking and molecular dynamics, to deliver comprehensive structural insights. This approach helps to fine-tune interactions at the binder-target interface, ensuring your biologic therapeutics are both effective and safe, enhancing biologics drug discovery.
Explore unique blend of AI technology and biological expertise
Advanced AI solutions for biologics discovery
Best-in-class tools for analyzing protein sequences and structures enable rapid identification of the most promising candidates.
Comprehensive knowledge and insight
Proficiency in AI, bioinformatics, and physics-based techniques ensures a thorough understanding of protein structures and interactions, critical for optimizing biologics’ therapeutic potential.
Proven success:
With a history of delivering impactful results across various projects, our solutions have consistently led to improved outcomes, including enhanced binding affinities, higher production yields, and minimized off-target effects.
See also:
Contact
Ready to transform drug discovery?
Discover how one of the top AI CROs in the world, can be your trusted partner in revolutionizing drug discovery through AI.
Contact us today to learn more about our tailored solutions for empowering your drug development journey.
Send us a message and we will contact you back within 48 hours.
Newsletter
Become an insider
Be the first to know about Ardigen’s latest news and get access to our publications, webinars and more!