Antibody Hit Screening: Developing Successful Therapies with the Help of In Silico Tools
Artificial intelligence (AI) is transforming drug discovery and development, including antibody-based therapeutics. This blog post explains how in silico tools can help find the best candidates for therapeutic antibodies from the pool of potential candidates. To learn more about advanced computational methods for the development of antibody-based therapies, check out the previous AI-powered Breakthroughs in Antibody Lead Optimization blog post.
Table of Contents:
- Computational approaches to antibody screening
- Developability
- Liabilities
- Combining different types of in silico tools for best results
- Ready to develop successful antibody-based therapies?
In developing antibodies, the screening stage plays a crucial role in narrowing down the pool of candidates to the ones with the best chances of becoming successful therapies.
Traditionally, antibody screening had been done in vitro using different assays. This involves screening tens and hundreds of thousands of molecules, resulting in high costs and long development times. Today, researchers can use in silico screening tools, including ‘traditional’ bioinformatics tools as well as AI-based methods for predicting antibody properties and selecting the most promising candidates.
Computational approaches to antibody screening
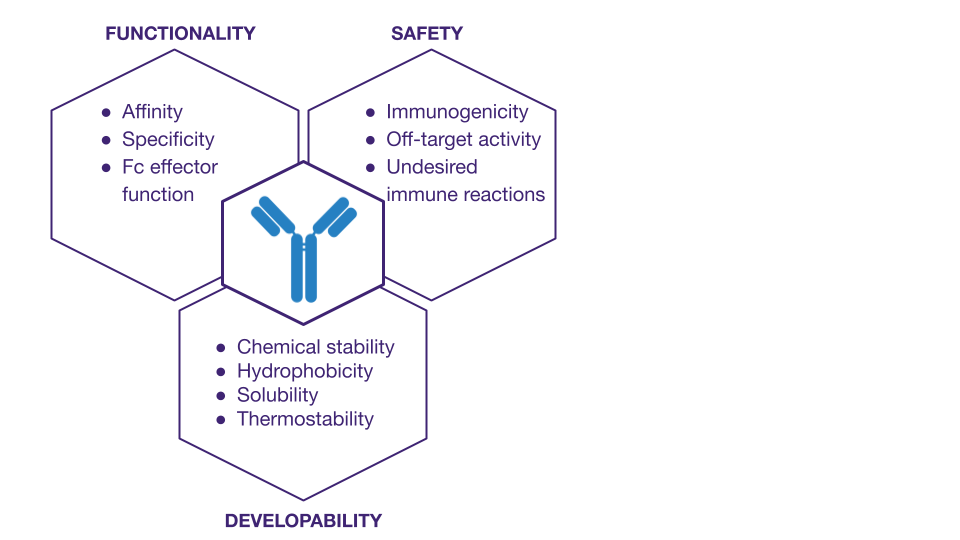
Computational analysis tools reduce the need for in vitro screening by using in silico filtering methods to pre-select candidates that have the highest likelihood of being developed into successful therapies. Typically, hit screening involves analyzing and optimizing for multiple parameters, such as:
- functionality (affinity, specificity, or Fc effector function)
- safety (immunogenicity, off-target activity, or undesired immune reactions such as Anti-Drug Antibodies or ADAs)
- developability (chemical stability, hydrophobicity, solubility, aggregation tendency, and thermostability)
While functionality is undoubtedly of great importance when screening antibody candidates, we are going to focus primarily on developability, liabilities, and safety.
Developability
‘Developability’ is a broad term that describes the feasibility of successful antibody development, which can be assessed by analyzing the candidate’s physicochemical properties [1]. Antibody developability includes factors such as solubility, stability, manufacturability, and storability. Developability characterization is key for preventing downstream issues that can lead to delays, higher development costs, or even failure.
Sequence-based tools, such SoluProt, a bioinformatics tool for solubility prediction, can be used to identify sequence liabilities mentioned above [2]. Another sequence-based tool called BioPhi can be used for immunogenicity prediction and humanization [3].
Structure-activity relationships (SAR) approaches can be used to predict the developability of an antibody based on its structure. The structure can either be obtained experimentally or predicted by AI models like AlphaFold or its derivatives. An example of structure prediction tools specialized in antibodies is the ABodyBuilder3 a recently released tool from Charlotte Deane’s group. Structure-based tools include FreeSASA, a model for calculating solvent-accessible surface areas [4], and PROPKA for estimating electrochemical properties such as isoelectric point and charge heterogeneity and 3D-based [5, 6].
A more comprehensive assessment can be done using the Therapeutic Antibody Profiler (TAP), a sequence- and structure-based computational tool for antibody screening. TAP compares the 3D biophysical properties of antibody candidates to those of clinical-stage therapeutic antibodies [7] and enables high-throughput scoring of antibodies based on developability factors such as the length of the CDRs, hydrophobicity, and the presence of charge patches that can link to aggregation [8].
However, using TAP as a webserver tool is not ideal for commercial purposes, as it may present challenges when employed as a high-throughput solution and poses risks due to the sensitive nature of antibody sequences as valuable assets. To ensure data security, it is safer for companies to run such analyses on their premises. At Ardigen, we specialize in developing tools that can be deployed on client premises, ensuring compliance and safeguarding critical data.
Liabilities
Antibody sequence liabilities are sequence motifs that can negatively affect antibody expression, conformational stability and ability to bind antigens. These include premature stop codons or frameshift mutations, post-translational modifications (PTMs), deamidation, oxidation and isomerization that could cause misfolding or aggregation. For example, PTMs in complementarity-determining regions (CDRs) of an antibody reduce its potency [9].
Among the most common sequence liabilities are including amino acid repeats and hydrophobic patches, which can negatively affect antibody folding and solubility. Liabilities can be generally detected through sequence analysis using tools such as Post-translational Modification Enrichment Integration and Matching Analysis (PEIMAN).
Safety
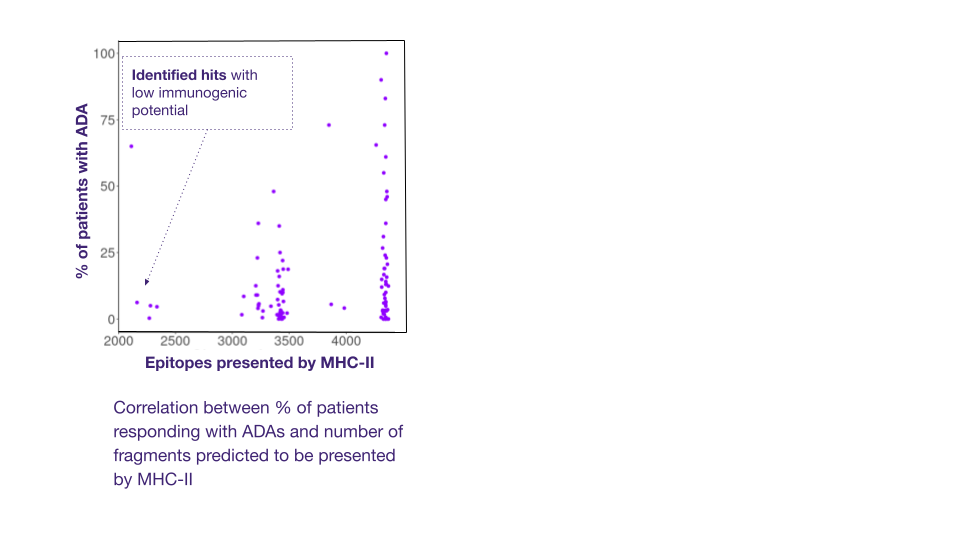
Immunogenicity is a serious problem for antibody therapeutics. Adverse immunological reactions to biologics, such as the development of Anti-Drug Antibodies (ADAs), can cause changes in toxicology, pharmacokinetics, and efficacy, and result in serious allergic reactions, including anaphylactic shock [10]. Some of the approved antibodies showed adverse effects in more than 70% of patients [11].
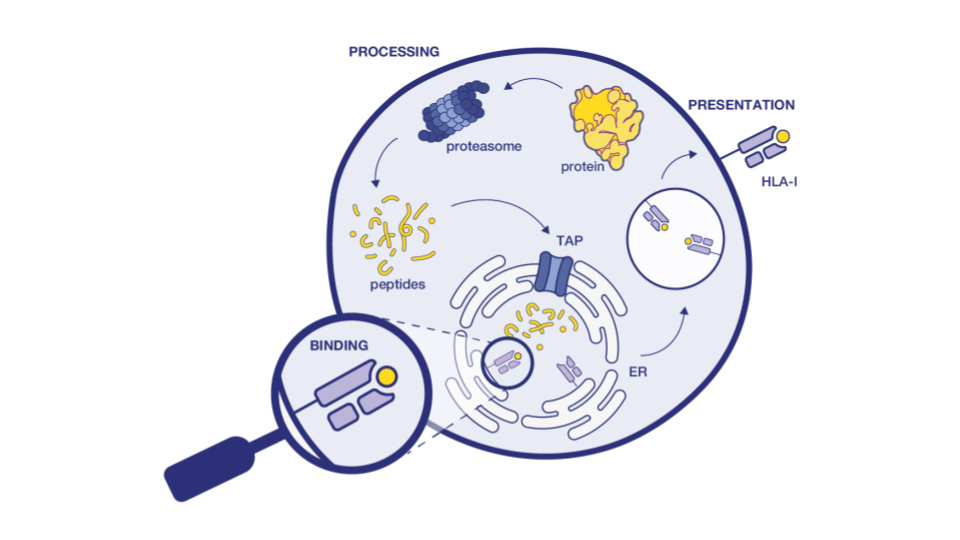
ADAs detection methods are essential during antibody hit screening. These rely on the detection of T cell epitopes in the primary sequences of antibody-based drugs, which are the key drivers of ADA response. In silico tools can identify the presence of potential T cell epitopes in the protein. Ardigen has a best-in-class model, ARDisplay-II, for the prediction of peptide presentation by HLA-II molecules, which significantly facilitates the screening of antibody drug candidates. When benchmarked against other models like netMHCpan 4.1 (EL) and MHCflurry (EL), our models demonstrated 2x higher predictive performance for eluted ligand (EL) solutions.
Combining different types of in silico tools for best results
The main challenge for AI-based tools is the limited availability of open-source, antibody-specific data needed to train these models. For instance, the SSH2.0 model was strained on a dataset from a 2017 study [12] which compiled the sequences and experimental assay results of 137 antibodies that had gained approval or had reached Phase 2 or Phase 3 clinical trials. To date, this remains the largest publicly available experimental data training dataset for developability assessment.
Ardigen is tackling this challenge by using general Large Language Models (LLMs) for proteins. LLMs are trained on a vast amount of data from both public and proprietary protein datasets to obtain models that capture general protein design rules. These models can be subsequently fine-tuned with application-specific data from our partners and clients that captures the properties of interest, such as antibody developability. The ideal basis for such projects is PRISM, Ardigen’s tool that combines several protein large language models (LLMs) that allow for the development of more specific models enhanced with partners’ data.
Our bioinformatic analysis pipeline enables the identification of sequence liabilities by analyzing properties such as the length of CDR chains, surface hydrophobicity, positive or negative charge patches in CDR proximity, structural charge symmetry in Fv region, and other parameters. We also use protein sequence and structure-based analysis as well as perform a comparison of the candidate properties with those of known clinical-stage therapeutics.
Ready to develop successful antibody-based therapies?
When it comes to antibody hit screening, using in silico tools to pre-select the candidates with the highest likelihood of success can save a lot of time and resources down the road. Computational tools can help ensure that the candidates you select have good developability properties, whether you are starting from protein sequences, structures, or functionalities. Investing in hit screening up-front will help you develop successful antibody-based therapies in a fraction of the time.
At Ardigen, we use best-in-class bioinformatics and AI-based methods for predicting antibody properties. With AI prediction tools, we can filter candidates using existing pre-trained models as well as create custom AI models powered by client-provided data. Reach out to us to learn how you can leverage in silico hit screening tools to increase the success rate of your antibody-based therapies.
Are you interested in biologics and would like more details? Get in touch!
Works Cited:
[1] Zhang, W. et al. Developability assessment at early-stage discovery to enable development of antibody-derived therapeutics. Antib. Ther. 6, 13–29 (2023). https://academic.oup.com/abt/article/6/1/13/6823522
[2] [Hon, J. et al. SoluProt: prediction of soluble protein expression in Escherichia coli. Bioinformatics 37, 23–28 (2021). https://academic.oup.com/bioinformatics/article/37/1/23/6070085
[3] Prihoda, D. et al. BioPhi: A platform for antibody design, humanization, and humanness evaluation based on natural antibody repertoires and deep learning. MAbs 14, (2022). https://www.tandfonline.com/doi/full/10.1080/19420862.2021.2020203
[4] Mitternacht, S. FreeSASA: An open source C library for solvent accessible surface area calculations. F1000Res. 5, 189 (2016). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4776673/
[5] Søndergaard, C. R., Olsson, M. H. M., Rostkowski, M.& Jensen, J. H. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. J. Chem. TheoryComput. 7, 2284–2295 (2011). https://pubs.acs.org/doi/abs/10.1021/CT200133Yb
[6] Olsson, M. H. M., Søndergaard, C. R., Rostkowski, M.& Jensen, J. H. PROPKA3: Consistent Treatment of Internal and SurfaceResidues in Empirical pKa Predictions. J. Chem. Theory Comput. 7, 525–537(2011). https://pubs.acs.org/doi/abs/10.1021/ct100578z
[7] Raybould, Matthew IJ, et al. “Five computational developability guidelines for therapeutic antibody profiling.” Proceedings of the National Academy of Sciences 116.10 (2019): 4025-4030. https://www.pnas.org/doi/abs/10.1073/pnas.1810576116
[8] Raybould, Matthew IJ, et al. “Contextualising the developability risk of antibodies with lambda light chains using enhanced therapeutic antibody profiling.” Communications Biology 7.1 (2024): 62. https://www.nature.com/articles/s42003-023-05744-8]
[9] Bailly, Marc, et al. “Predicting antibody developability profiles through early stage discovery screening.” MAbs. Vol. 12. No. 1. Taylor & Francis, 2020. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7153844/
[10] Pratt, Kathleen P. “Anti-drug antibodies: emerging approaches to predict, reduce or reverse biotherapeutic immunogenicity.” Antibodies 7.2 (2018): 19. https://www.mdpi.com/2073-4468/7/2/19
[11] Vaisman-Mentesh, Anna, et al. “The molecular mechanisms that underlie the immune biology of anti-drug antibody formation following treatment with monoclonal antibodies.” Frontiers in immunology 11 (2020): 1951. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7461797/
[12] Jain, Tushar, et al. “Biophysical properties of the clinical-stage antibody landscape.” PNAS 114.5 (2017): 944-949. https://www.pnas.org/doi/abs/10.1073/pnas.1616408114




