In today’s life sciences landscape, data isn’t just growing – it’s exploding. Sequencing is cheaper than ever (see the NHGRI data). Multi-omics platforms: spanning genomics, transcriptomics, proteomics, and metabolomics, are generating complex datasets at unprecedented speed and scale (view market growth trends). This should be a golden age for biomedical insight.
But here’s the catch: turning raw data into meaningful insight, especially at scale, isn’t just a matter of storage or access. It’s about ensuring your tools, workflows, and infrastructure are built to handle the volume, variety, and velocity of modern biomedical data without falling apart. Building robust drug discovery data infrastructure is the key to harnessing petabyte-scale datasets and this post explores how.”
It’s Not Just Volume. It’s Complexity. And Speed.
Let’s say a team builds a clever algorithm that flags new therapeutic targets based on multi-omics input. It works beautifully on clean, curated datasets, maybe even outperforms expectations.
But once that same algorithm is applied to real-world clinical data – thousands of patients, dozens of omics layers, missing values, inconsistent formats – it breaks. Processing slows to a crawl. Cloud costs balloon. What seemed like a scalable solution quickly turns into a bottleneck.
And it’s not just the amount of data – though it often reaches petabyte scale. The real challenge lies in the velocity at which data is generated, and the variety of formats, sources, and contexts. Integrating DNA sequences, RNA expression levels, protein interactions, and clinical records introduces multiple layers of complexity (see diagram of integration strategies) that most systems aren’t designed to handle efficiently.
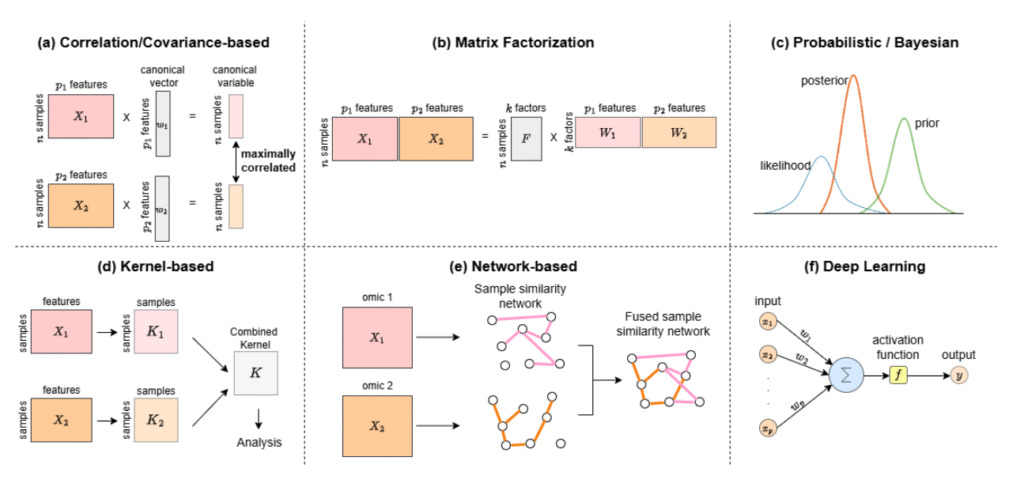
Figure adapted from: Baião, A.R., Cai, Z., Poulos, R.C., Robinson, P.J., Reddel, R.R., Zhong, Q., Vinga, S., & Gonçalves, E. (2024). A technical review of multi-omics data integration methods: from classical statistical to deep generative approaches. INESC-ID, Instituto Superior Técnico (Universidade de Lisboa) & ProCan®, The University of Sydney. https://doi.org/10.48550/arXiv.2501.17729
It’s a common trap. Most bioinformatics tools were developed in academic settings, meant to explore interesting biological questions, not to survive enterprise-grade deployment. That’s where things start to fall apart:
- Algorithms don’t scale.
- Data pipelines aren’t built for automation or reproducibility.
- Integration across formats becomes a headache (see example integration strategies).
- Cloud resource usage spirals out of control (understand cloud cost challenges).
Hardware Acceleration as a Turning Point
Once data pipelines hit their scaling limit, small optimizations aren’t enough. You need a fundamental shift in how work gets done. One of the most impactful enablers? Hardware acceleration, especially with GPUs.
Originally designed for graphics rendering, GPUs are now central to high-performance computing in life sciences. Their architecture -thousands of cores handling parallel tasks, is a perfect match for the kinds of problems bioinformatics throws at them.
The gains aren’t theoretical. They’re measurable:
- Genomic variant calling used to take days on CPUs. Now, tools like Illumina’s DRAGEN Bio-IT Platform and NVIDIA Parabricks compress that into hours or even minutes without compromising accuracy.
- Single-cell RNA-seq, notoriously heavy on compute, benefits enormously from GPU acceleration. Traditional tools like scanpy can become painfully slow at scale. But frameworks like RAPIDS and rapids-singlecell routinely offer 10–75× speedups. That’s not just faster results—it enables researchers to ask new kinds of questions and run more iterative, exploratory workflows.
- AI and ML applications in drug discovery thrive with faster processing. With GPU acceleration, teams can build deeper models, run more frequent training cycles, and extract subtle patterns from omics data that would be missed otherwise (read more about AI’s impact).
The key takeaway? It’s not just about doing the same work faster. It’s about doing more meaningful work. Work that wasn’t previously feasible.
Speed Is Nothing Without Structure
All the acceleration in the world won’t help if your data is scattered, messy, or poorly documented. Without a solid foundation, even the most powerful tools will struggle to deliver.
That’s why the FAIR principles (Findable, Accessible, Interoperable, Reusable) are no longer just a best practice. They’re essential. They create a framework for managing data in a way that supports collaboration, auditability, and reuse across teams and tools (learn more about FAIR).
Strong foundations also require the right people. Teams that blend scientific insight with technical depth—bioinformaticians, computational biologists, software engineers, and IT architects—are the ones that tend to build systems that scale.
Read our blog on taming data chaos in drug pipelines
From Concept to Production
Going from an idea to a robust, scalable pipeline isn’t a straight line. It takes strategy, iteration, and real-world experience.
Some lessons we’ve learned along the way:
- Design pipelines that are modular, portable, and transparent.
- Pick tools that scale—not just ones that are familiar.
- Plan for cloud from the beginning. Use managed services and cost controls wisely.
- Automate as early as possible. Tools like Nextflow, Snakemake, and Cromwell make reproducibility and orchestration easier.
- Monitor cost and performance continuously—not just at launch.
Partnering with experienced teams who understand both omics and compute infrastructure can be the difference between a stalled prototype and a production-ready system.
Join the Conversation
If you’re exploring how to scale your omics workflows, reduce bottlenecks, or integrate hardware acceleration into your infrastructure, we invite you to our upcoming webinar:
“Turning bioinformatics concepts into production engines through GPU acceleration”
June 24, 2025 | 5 PM CEST
We’ll cover:
- Common challenges that prevent bioinformatics projects from scaling
- How tools like Parabricks and RAPIDS can change what your team can deliver
- What it takes to move from idea to production in real-world settings
- Examples of successful transformations
Don’t let data bottlenecks stall your innovation. Learn how to transform your computational challenges into strategic advantages. We look forward to sharing our insights and helping you navigate the future of biomedical data processing!
Need help scaling your drug discovery pipelines? Visit our Knowledge Hub or contact us for a consultation.




