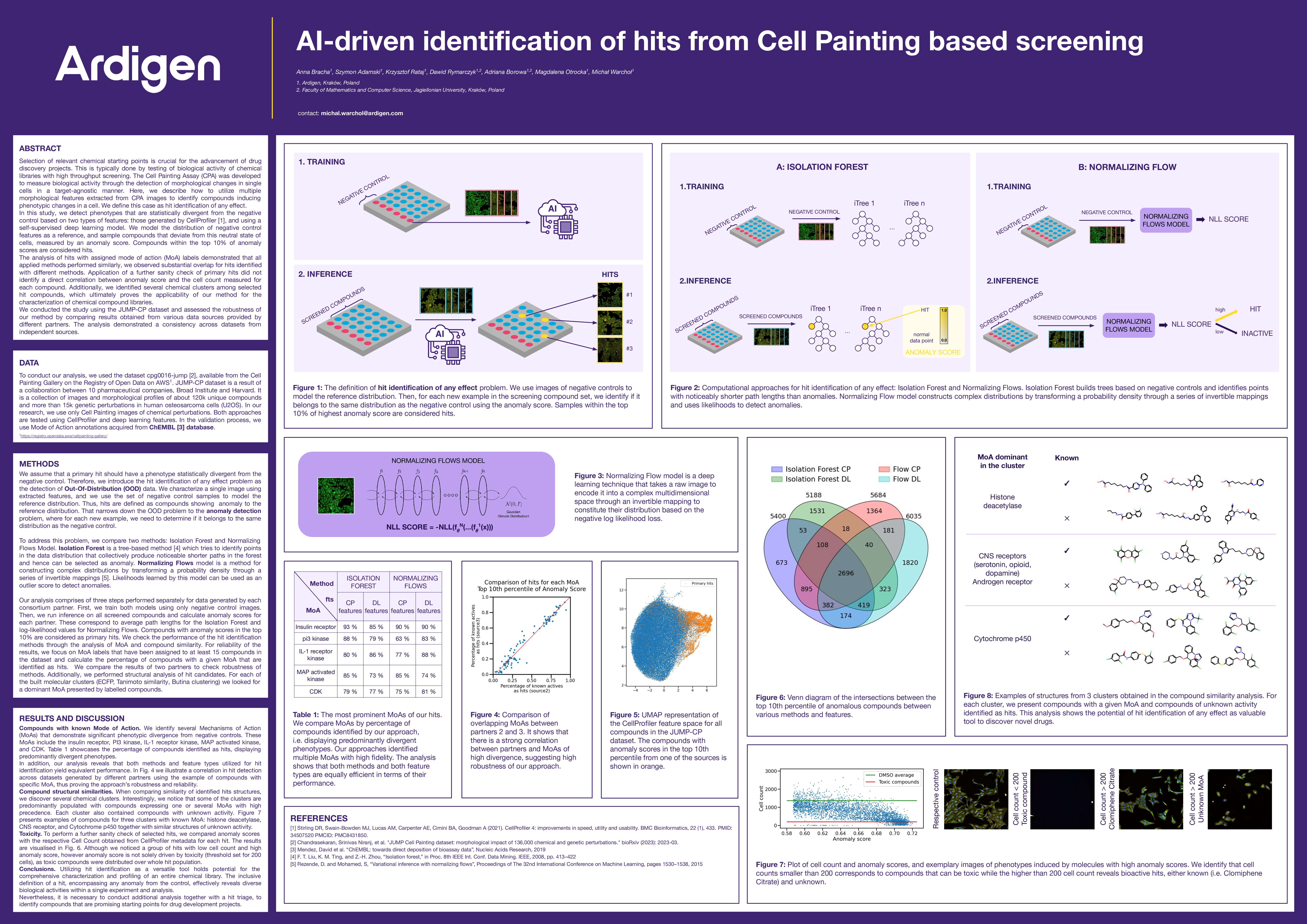
At Ardigen, we continuously seek innovative approaches to decode complex biological systems. At the recent SBI2 2023 conference, our team presented a poster showcasing an AI methodology for identifying active compounds in high-content phenotypic screening. Using Cell Painting Assays (CPA) and cutting-edge anomaly detection models, we demonstrate how artificial intelligence can significantly enhance hit identification—capturing subtle cellular responses that might otherwise go unnoticed.
Understanding the challenge: Hit Identification of any effect
High Content Screening, such as the Cell Painting Assay, captures rich morphological profiles of cells treated with thousands of chemical compounds. But within this ocean of data, how do we distinguish truly bioactive compounds – those inducing meaningful cellular changes – from noise?
Our answer lies in reframing the problem: we don’t search for known effects, but for any effect—that is, any phenotype statistically divergent from the baseline. This turns the challenge into a problem of anomaly detection.
Our approach: Merging AI and cell morphology
We evaluated two computational approaches for detecting out-of-distribution (OOD) phenotypes:
- Isolation Forest – a tree-based method that identifies anomalies by how isolated data points are in feature space.
- Normalizing Flows – a deep learning-based method that models complex probability distributions, allowing us to assess how likely (or unlikely) a given cell profile is under the control distribution.
These models were trained using only negative control samples, then used to score all other compounds. The top 10% with the highest anomaly scores were flagged as potential hits.
We applied this methodology to the JUMP-CP dataset—a collaboration of 10 pharmaceutical companies and academic partners, comprising over 120,000 chemical perturbations. Features were extracted both via traditional tools like CellProfiler and modern self-supervised deep learning models.
Key findings: More hits, greater chemical diversity
- Equivalent performance across models and features
Both models—despite differing in complexity—consistently identified similar sets of hits across partners. This suggests robustness and reproducibility of our approach. - Meaningful MoAs emerge from anomalies
Among identified hits, many belonged to known mechanisms of action (MoAs), including insulin receptor, PI3 kinase, MAP kinase, and more. These phenotypically divergent groups confirmed the biological relevance of our anomaly scores. - Uncovering structural clusters
We clustered hit compounds based on chemical similarity and found distinct groups sharing MoAs, alongside unexplored compounds of unknown activity. This highlights the potential of our method to surface novel leads for drug discovery. - Toxicity not the whole story
A cross-check with cell count revealed that high anomaly scores weren’t solely due to cytotoxicity. Many bioactive hits maintained healthy cell counts, underscoring the method’s ability to detect nuanced, non-lethal phenotypic effects.
Conclusion: From screening to strategy
Our approach illustrates how AI anomaly detection in Cell Painting can be a powerful, agnostic method to probe the functional space of chemical libraries. By expanding the definition of a “hit” to include any significant phenotypic shift, researchers gain a more inclusive and potentially transformative tool for early drug discovery.
While this is a foundational step, further triage and validation remain key. Yet the prospects are exciting: marrying rich imaging data with modern machine learning allows us to chart new territory in phenotypic profiling—one outlier at a time.