In the biomedical world, a vast amount of diagnoses, clinical trials, drug development, and decisions are made daily, based on collected data. In many of these processes, the image plays either the primary or supporting role. But what is this image? What kind of information is it carrying? How much of it is hidden in there? And are we, human beings, able to read them thoroughly when they are acquired in a laboratory environment, vastly different from our natural one? How should we analyze these images?
In recent years, there have been many breakthroughs in computer vision. In some tasks, the algorithms can perform on par or even outperform humans. Achieving such performance is due to the accessibility of enormous databases, powerful machine graphical processors, and researchers, striving to develop and enhance the deep learning techniques.
A plurality of biomedical image types
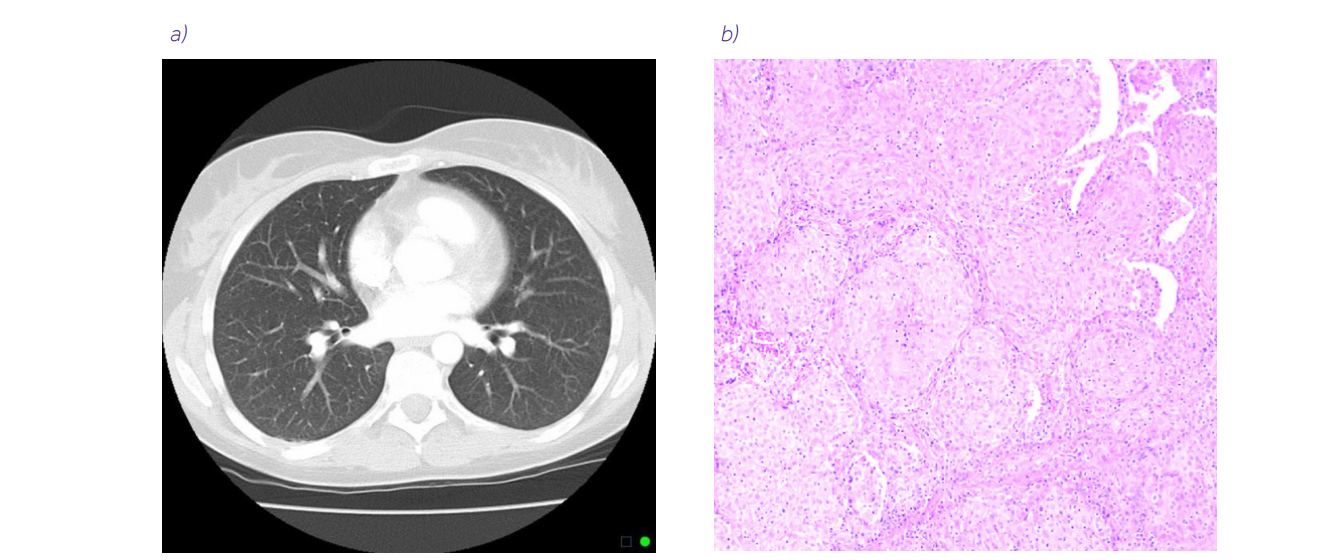
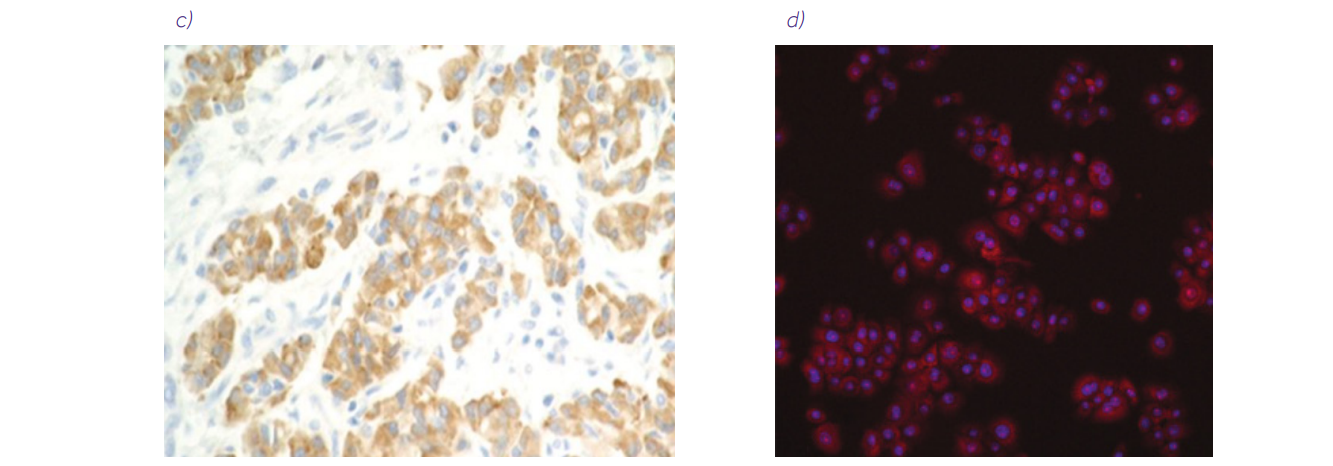
Before we dive into machine learning and artificial intelligence, we need to understand how different biomedical images are in contrast to natural ones. Databases are limited, and collecting the data, especially in clinical trials, is expensive and tedious, because it requires highly-experienced staff. While one of the most common natural image databases, ImageNet, consists of more than 14 million images of 1000 classes, the biomedical databases consist of hundreds of samples, like in PANDA challenge (11 000), Camelyon17 (899), or LIDC-IDRI (1018). These archives are for common diseases, like breast, prostate, or lung cancer, but the rarer the disease, the smaller the dataset is. Moreover, biomedical images have different modalities, like CT, MRI, PET, USG, histology, and other types of scans. And that is not all – e.g. in the field of histology, researchers are using H&E stained or immunochemistry images, in High Content Screening, we have scans in light of different wavelengths, and in MRI, we have T1 and T2 views or even a stack of the images from which we can reconstruct an organ. Figure 1 presents examples of biomedical images of lungs.

Figure 1. Examples of biomedical images of lungs. a) Normal chest CT scan source. b) Lung biopsy slide with hematoxylin and eosin staining (10× magnification) source: An 11-Year-Old Boy with Diffuse Pulmonary Infiltrates and Respiratory Failure – Scientific Figure on ResearchGate. c) Immunochemistry biopsy slide (source: 10.1183/16000617.0007-2017) from the article Nonsmall cell lung carcinoma: diagnostic difficulties in small biopsies and cytological specimens, used under CC BY 4.0 d) Cells prepared for high content screening (source: 10.3791/53987) from the article High Content Screening Analysis to Evaluate the Toxicological Effects of Harmful and Potentially Harmful Constituents (HPHC), used under CC BY-NC-ND 3.0
Having in mind such variation in the images does not make it possible to create one unified analysis module. That is why at Ardigen, we have developed a technology platform that can master them all. Through our expertise, we can choose the best possible computer vision approach to analyze a certain image type and address challenges posed by it. For histological images, we need to deal with differences in staining and tissue preparation between the medical centers. The microarray image analysis presents a challenge because data comes from scanners and imagers, along with their artifacts, and then HCS images can be presented in a 3D form or 2D slice.Challenges in biomedical image analysis
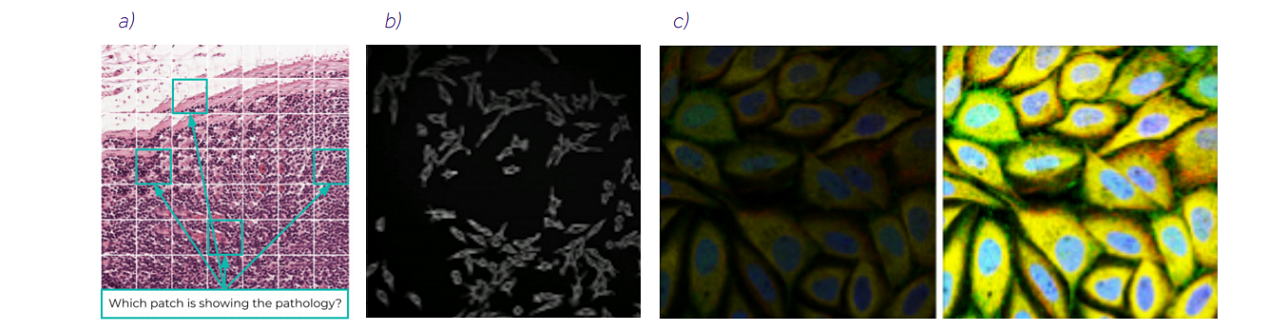
Before we dive into the details about the image analysis, let us introduce few challenges that we encountered during our projects: • image of super-resolution: to properly show a single cell or a spot, many of the images, like HCS, histology, or microarray data, need to be acquired with high magnification. Depending on the task, the images can be resized (e.g. microarrays to register the grid) or partitioned into smaller patches (e.g. HCS, histology) to preserve a piece of valuable information, • weak labels, which do not directly correspond to the single data point (e.g. patch), but the bag of the instances (e.g. set of patches made from whole-slide biopsy images), • batch effects, which can be related to the device acquiring the image (imager vs scanner) or to different laboratories the make the image and prepare the materials, • data imbalance, which underrepresents the edge cases in the dataset, because they are not common in the population, • data quality: even curated databases have issues that can influence the AI algorithms, especially datasets which were not intended to be used with AI, like blurriness, • labels’ quality, because there can be a discrepancy between doctors’ diagnoses or human annotators, • relatively small number of images available for algorithm training, compared to natural image datasets like ImageNet, • artifacts related to the devices, like luminance glitches, distortions, or any other image defects, • presenting the rationale behind the prediction, which is essential when the algorithm is presenting the decision that may have the human life at stake, • assessment of the reliability of the prediction: whether it is trustworthy or not, • and many, many more.
Figure 2. a) Example of weak label. We know the image presents pathology, but we do not know which part it is. b) Example of data quality issue: blurriness. c) Example of batch effects, images present the same data, but were taken with different light strength.
Succeeding the biomedical image analysis
The key to successfully overcoming these challenges is the cooperation of the engineers, researchers, and daily practitioners in order to build a system suited for a particular task. We frame such teams for each of the projects that we are working on to maximally substantiate the success. The process of building the image analysis pipeline is based on our software components that speed up the delivery and cooperation to achieve a scalable, reliable, and trustworthy system. More details on our projects, components, solutions to the challenges and results will be presented in future blog posts.Application of biomedical image analysis
Now, we will point out some interesting applications of computer vision algorithm to the analysis of the biomedical images to show the potential that is hidden in them: • Repurposing the drugs using the High-Content Screening images, • Finding the gene mutation, based on the H&E stained whole-slide biopsy images, • Classifying the fungi specie based on the microscopy image and not the biochemical test, • Detecting the breast cancer on mammography images with accuracy on par with those of medical experts.Bibliography
- He, Kaiming, et al. “Delving deep into rectifiers: Surpassing human-level performance on imagenet classification.” Proceedings of the IEEE international conference on computer vision. 2015.
- Deng, Jia, et al. “Imagenet: A large-scale hierarchical image database.” 2009 IEEE conference on computer vision and pattern recognition. IEEE, 2009.
- https://www.kaggle.com/c/prostate-cancer-grade-assessment
- https://camelyon17.grand-challenge.org/
- Armato III, Samuel G., et al. “The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans.” Medical physics 38.2 (2011): 915-931.