Examining the response of a cell to a certain treatment, disease, or phenomenon is the initial step for most biological analyses. Since the human body contains over 30 trillion cells and most of them contain nuclei with DNA, being able to rapidly and precisely localize the nuclei is crucial for a researcher to understand the biological processes. In this post we present a possible approach for automated nucleus detection. The algorithm is based on Mask R-CNN, a state-of-the-art deep learning method for object detection and detailed segmentation.
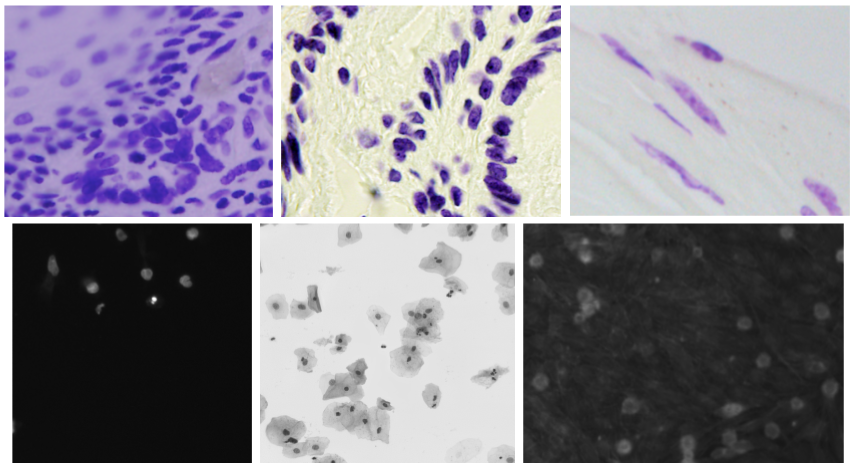
Image 1: Example data from competition dataset Source: kaggle.com
Ardigen AI Labs decided to participate in this competition; therefore, we formed a core team (Dawid Rymarczyk, Łukasz Maziarka, and Jan Kaczmarczyk, PhD), supported by the other data scientists from our company (Oleksandr Myronov, Bartosz Zieliński, PhD, Szymon Wojciechowski, and Michał Warchoł, PhD). In addition, we collaborated with the Academic Computer Center Cyfronet AGH for heavy GPU computations. In the beginning, we focused on reviewing the state-of-the-art methods, and we concluded that the competition challenge could be addressed in two ways—using instance segmentation with heavy post processing or semantic segmentation. After a discussion, we decided to apply a Mask R-CNN model for this task. The model was developed by Facebook AI Research in 2017, and since then, it is a state-of-the-art method in semantic segmentation. The Mask R-CNN model first generates a feature map from a backbone neural net, for example, Squeeze-and-Excitation ResNeXt (only convolutional layers are considered in the model because they provide a robust image representation). After extracting the feature map from the image, the model detects regions of interest (RoI), which are candidates for locations of objects (they are expressed in the form of coordinates of bounding boxes). In the next step, RoIs are filtered to remove those that overlap significantly, and then the remaining ones are classified as background or object. In the next step, each RoI goes through convolutional layers and at the end, there are three outputs: first for the bounding box of the object, second for the object’s class, and the third one for the segmentation. The segmentation output has fixed dimensions (e.g., 28 × 28 pixels) and therefore, needs to be resized to the original bounding box dimensions at the end of the pipeline.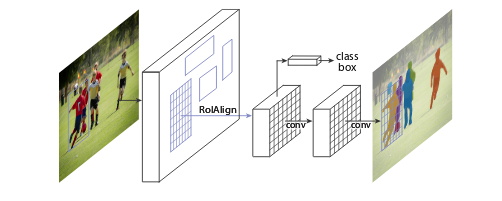
Figure 2: Mask R-CNN framework Source: https://arxiv.org/abs/1703.06870
We were aware that it is not enough to simply use the standard Mask R-CNN method to achieve a prize or even a decent rank; therefore, we started to precisely analyze the input data. We followed discussions on related forums, analyzed Mask R-CNN follow-up papers, and we followed the guidelines from the organizers to focus on data heterogeneity and the importance of micronuclei. The first improvement we introduced in the baseline model was data normalization technique called CLAHE. Unfortunately, it did not preserve information on color contrast on the same intensity level. Therefore, as an alternative, we decided to add an additional channel H (from HSV image representation) into the input. Another very important step was to retrieve similar types of images from external medical databases. Including more images in the training dataset can be beneficial for obtaining proper image representation and improving model generalizability. To ensure robustness, data augmentation during the training phase is necessary. Therefore, we used the following transformations: rotation by 90°, 180°, and 270°; Poisson noise; vertical or horizontal flipping; warping; color perturbation; and focus or blur.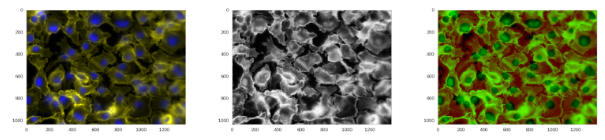
Figure 3: Original image, image normalized using CLAHE, and image normalized using our approach
When the input data for training was fully prepared, we were able to work more intensively on the algorithm itself. In a basic Mask R-CNN model, the ResNet model is used as a backbone, but we have decided to use a novel extension of it called Squeeze-and-Excitation ResNeXt, which won the ImageNet competition in 2017. Another enhancement was to replace the standard convolutional layers with dilated ones. Dilated convolution, introduced by Hamaguchi et al., focuses more attention on the broader context of the image, which should work better in case of micronuclei detection. We also used Cascade R-CNN architecture to provide a model dealing with different sizes of overlapping regions. Another addition to our model was the usage of Focal Loss function and usage of different mask sizes.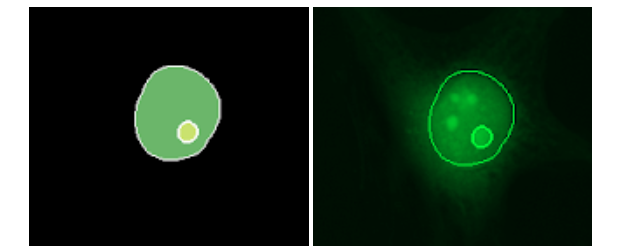
Figure 4: Segmentation and detected nucleus and micronucleus.
With the help of all these enhancements in the model, we could work on the post-processing steps. We applied two major techniques—filling holes and test-time augmentation. Test-time augmentation is making a prediction on original data and rotated/flipped data, aggregating them into one final prediction. As a result, we obtained a model that could successfully work with different types of data and detect numerous micronuclei. Training time on GeForce P40 required 92 hours. The final score of our team was 0.445, whereas the winning solution obtained a score of 0.631 and was based on U-Net. Finally, we secured the bronze medal and our solution was among 11% of the best in the world. Unfortunately, following the guidelines from the organizers did not guarantee winning the challenge. Moreover, we should have started with a simpler method (such as U-Net) and tried to explore it as much as possible, before getting into a considerably more complicated model (such as Mask R-CNN).Works Cited:
[1] He K., Gkioxari G., Dollar P., and Girshick R., Mask R-CNN, https://arxiv.org/abs/1703.06870
[2] Hu J., Shen J., and Sun G., Squeeze-and-Excitation Networks, https://arxiv.org/pdf/1709.01507
[3] Cai Z. and Vasconcelos N., Cascade R-CNN: Delving into High Quality Object Detection, https://arxiv.org/abs/1712.00726
[4] Lin T., Goyal P., Girshick R., He K and Dollár P., Focal Loss for Dense Object Detection, https://arxiv.org/abs/1708.02002