 If you were asked to imagine a scientist working hard to discover a new drug, what would the image be? Certainly, you’d think about a white-coat clad person in a laboratory, mixing and measuring chemicals with a pipette. While this is what happens in later stages of drug development, you might be surprised that the reality is somewhat more mundane: a scientist spends a lot of time sitting and reading, whether in an office or at home. Despite all the technological progress, reading is still the backbone of scientific discovery.
If you were asked to imagine a scientist working hard to discover a new drug, what would the image be? Certainly, you’d think about a white-coat clad person in a laboratory, mixing and measuring chemicals with a pipette. While this is what happens in later stages of drug development, you might be surprised that the reality is somewhat more mundane: a scientist spends a lot of time sitting and reading, whether in an office or at home. Despite all the technological progress, reading is still the backbone of scientific discovery.
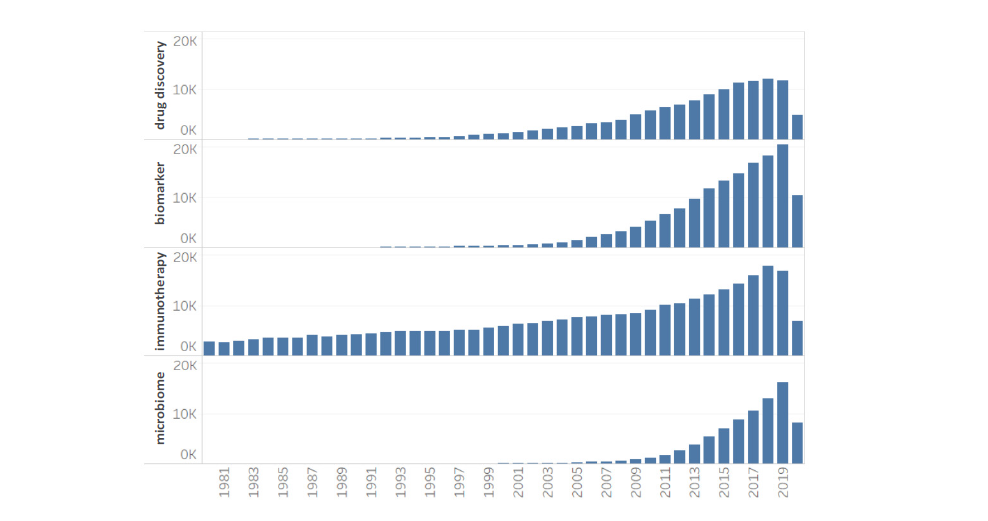
An average researcher spends about 15 hours per week reading and consumes about 250 articles every year. The thing is, even with reading as a daily habit, it is impossible to keep up with the incessant stream of content—articles, reports, and monographs—published every day. In fact, every year the total number of research papers in biomedical fields grows by 5%: in 2018 there were about 1.4 million new articles indexed in PubMed, while in 2019 the number went up to 1.47 million. Thus, a neuroscientist wishing to keep track of her field will have to dedicate more than two years just to read the 48,080 papers published in 2019. That is, if she never sleeps, eats, or does anything else than reading.
This begs the question: are we missing important opportunities because there is no way for humans to keep up with the knowledge we produce? Are we drowning in a sea of data?
A misstep in the early stages of the research process—reviewing literature—can have devastating logistical, financial, and scientific consequences: an overlooked breakthrough, a redundant clinical trial, or rediscovering what has been already established after much effort. The challenge to harness academic literature is especially daunting in the life sciences and medical research where the convergence of highly specialized disciplines leads to fragmented knowledge scattered across multiple domains.
Boosting research capabilities with AIKE
Scientists need help to cut through the complexity and volume of literature and recognise the hidden patterns that can lead to breakthroughs in medicine. At Ardigen, we can lend a hand. We have extensive experience in harnessing advanced AI algorithms to uncover the knowledge dormant within slews of data. We specialize in creating AI tools, capable of learning from different types of data, from genetic to metabolic information. Similar tools can be used to connect words and their contextual meaning within large literature repositories. By leveraging these algorithms we created an AI-powered Knowledge Extraction (AIKE) engine using Natural Language Processing (NLP) algorithms, to support our client’s drug discovery efforts. The engine is capable of extracting undiscovered knowledge from large collections of publications rapidly and effectively.
AIKE searches scientific publication repositories, such as PubMed, to catalogue and collect papers. These unstructured text data are then pre-processed to create a machine-readable text database (i.e. a corpus). Then, using NLP text-mining methods, AIKE searches and identifies connections between preselected concepts (Fig 2a). The engine can be operated from a user-friendly interface, which generates visualizations, e.g. to map out the knowledge dependencies. For example, AIKE can be used to connect brain regions with behavioural patterns or with drug targets and rank the publications supporting such connections in order of relevance (Fig. 2b).


Moreover, as scientific knowledge expands organically, AIKE keeps learning. We created a web-based application which scrapes scientific databases searching for newly published relevant articles to update the knowledge base. This way AIKE’s recommendations are up to date and relevant for cutting-edge research. While AIKE was developed for the study of neurobehavioral disorders, our engine can be repurposed for augmenting the discovery process in other research areas. In particular, to create an easy to grasp bird’s eye view of rapidly developing fields. This is especially valuable in times of urgent medical need, such as the COVID-19 pandemic. In the past six months, 18,436 papers on COVID-19 have been published. We believe that research augmentation tools such as AIKE will play an important role in this and future healthcare crises by making sure we do not miss anything important.
- Ware, Mark and Mabe, Michael, ”The STM Report: An overview of scientific and scholarly journal publishing” (2015). http://digitalcommons.unl.edu/scholcom/9
- Kimber Price, “Scientific literature overload: Tips for staying on top” (2018) https://scopeblog.stanford.edu/2018/08/28/scienti?c-literature-overload-tips-for-staying-on-top/
- https://pubmed.ncbi.nlm.nih.gov/?term=neuroscience&?lter=ds1.y_1&timeline=expanded
- Kirstin Borgerson, “Redundant, Secretive, and Isolated: When Are Clinical Trials Scientifically Valid?”(2014) https://pubmed.ncbi.nlm.nih.gov/25638948/
- https://pubmed.ncbi.nlm.nih.gov/?term=%22COVID-19%22




