About DREAM Challenges – Biology Experts United
NCI-CPTAC DREAM-Proteogenomics Challenge Through the Lenses of Ardigen Experts
The NCI-CPTAC DREAM-Proteogenomics Challenge [4] focused on predicting the protein concentrations in cancer tissues. Characterization and analyses of alterations in the proteome can shed more light on cancer biology and may improve the development of biomarkers and therapeutics. As measuring proteomic data is both difficult and expensive, the challenge objective was to develop machine learning models for predicting such information based on genomic and transcriptomic data.
The challenge consisted of three sub-challenges. Ardigen’s team took part in the second and third, securing the fifth and second place, respectively.
› For sub-challenge 2, their objective was to predict protein abundances based on genomic – that is, copy number alterations and CNA – and transcriptomic – meaning mRNA expressions.
› Sub-challenge 3 included both these goals with the addition of phosphoprotein data.
Our team’s solution applied directly to the sub-challenge 2. Therefore, now we will dive into its details while highlighting this segment.
About Proteogenomics
Proteogenomics is a multi-omics research approach that sheds new light on cancer development. Integrating genomic, transcriptomic, and proteomic information can lead to a deeper understanding of the complex processes related to cancer and the development of novel biomarkers and therapeutics.
NCI-CPTAC Dream Proteogenomics Challenge – the Data and the Task
The challenge data provided by the Clinical Proteomic Tumor Analysis Consortium (CPTAC) [5] measured for ovarian and breast cancer. The proteomic data was measured by two laboratories: John Hopkins University (JHU) and Pacific Northwest National Lab (PNNL) [6]. The median Pearson correlation of paired protein measurements from the same sample, measured in the two laboratories, was 0.69. This result already shows how difficult it is to measure the proteome.
The challenge dataset was high-dimensional with around 100 input observations – in other words, samples – and typically from 10,000 to 35,000 features. These include:
- CNAs,
- gene expressions,
- protein abundances [4].
In the 2 and 3 sub-challenges, the task was to predict around 1,000-15,000 target variables. The metrics used to measure the performance was the average Pearson correlation between predicted and measured (phospho)protein abundances.
The Ardigen Team Composition
The Ardigen’s team consisted of four experts who repeatedly discussed the model development strategy.
› Jan Kaczmarczyk proposed the general architecture of the models and implemented them,
› Łukasz Maziarka suggested vital improvements in the models,
› Piotr Stępniak provided bioinformatics support,
› Michał Warchoł guided model design.
NCI-CPTAC DREAM-Proteogenomics Challenge – the First Stage of Model Development
Dream Challenges – Ardigen’s Second Stage of Model Development
- 0.46 (for ovarian cancer) and 0.42 (for breast cancer) in sub-challenge 2,
- 0.29 (for ovarian cancer) and 0.36 (for breast cancer) in sub-challenge 3.
Our Model Architecture
During the challenge, we used a two-stage hierarchical forward feature selection algorithm. The first stage focuses on variables showing good generalization properties as determined by an auxiliary model. We used its residuals in the second stage by fitting them into a separate model.Model Architecture: Stage One
In stage 1, we choose the single, ‘most promising’ model among models built using linear regression on the following sets of 1-2 variables: mRNA, CNA (mRNA, CNA), hc: TFs, hc: all. Here, mRNA and CNA denote the features relating to the considered protein (i.e. with the same GeneID), the only set with two variables is (mRNA and CNA), and by hc, we mean the variable with the highest correlation to the target variable from a given set of variables. In particular hc: TFs denotes the variable chosen among mRNA and CNA of human transcription factors (TFs) [9], whereas hc: all variables denotes the variable chosen among all variables. From the hc: TFs and hc: all candidate feature sets we remove mRNA and CNA relating to the considered protein. We use TFs as a separate set of variables because limiting ourselves to a relevant subset of the original features should decrease noise.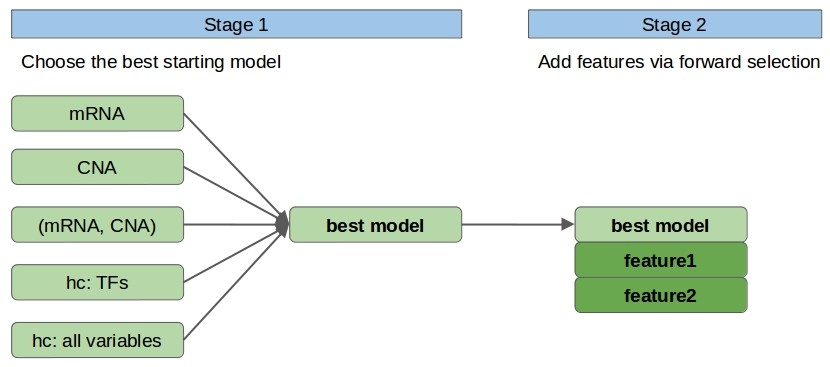
- Fig. 1. Model architecture
Model Architecture: Stage Two
In stage 2, we fit a separate model to the residuals of the model obtained in stage 1. To this end, we use a forward feature selection procedure. For the early-stopping criterion, we consider the cross-validation (CV) score change when adding a variable. We follow a customized cross-validation procedure with five folds and a holdout set. The holdout set (with ~20% of data) is used to obtain a more realistic (not overfit) estimation of model performance. Additionally, we impose a barrier, b, for the enlargement of the model by variable(s) being added. If the considered variable(s) does not improve the CV score by at least b = 0.06 (for ovarian cancer) or b = 0.07 (for breast cancer) it is rejected and the procedure stops. The values of b = 0.06, 0.07 were determined by optimizing for the holdout set score. Note that the feature with the highest correlation to the residuals of the current model is chosen per train-test split in the cross-validation procedure and can even be a different feature for each split.Choosing the Best Starting Model
-
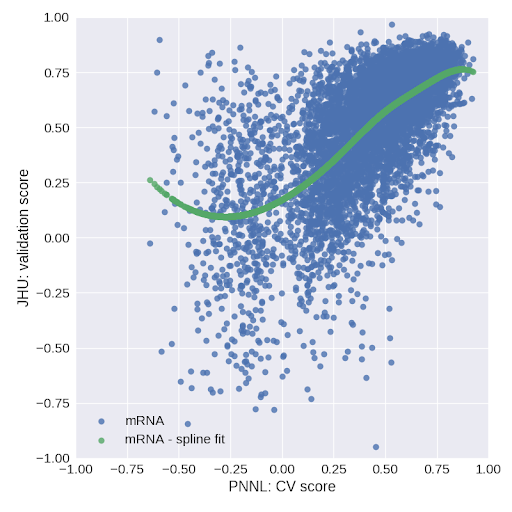
Fig. 2. Validation score on JHU data versus cross-validation score on PNNL data for stage 1 models trained on mRNA.
-
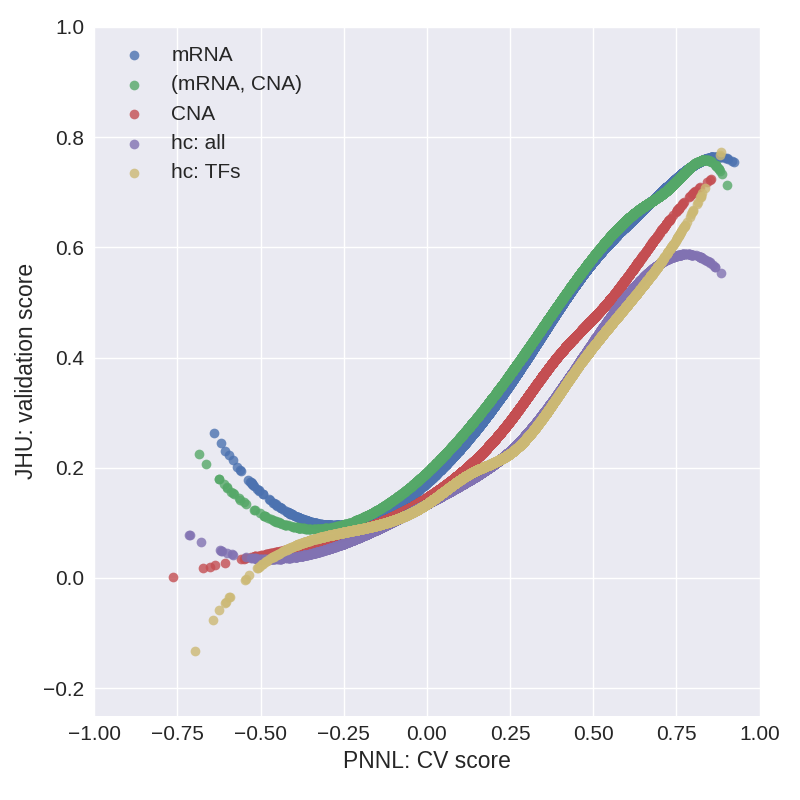
Fig. 3. Cubic spline fits to the (CV score, validation score) data for models considered in stage 1.
Sub-Challenges Model Details: Transformations of Variables and Ensembling
For all our models in sub-challenge 2 we use unregularized linear regression, whereas in sub-challenge 3 we use least angle regression (LARS) implemented in the LarsCV class in the sklearn package in Python.
For imputation we use the mean imputation. We also tested several transformations on the predictors, of which X→log(X+B ) and (X, Y )→(X, 2Y ) are included in our final ensemble of models. As a final solution, we use ensemble of models over different fold splits and – in sub-challenge 2 – with the above transformations of variables.
In our final submission, we use an ensemble of 15-65 models per (phospho)protein for both the ovarian and breast cancer datasets. The models within an ensemble differ by the splitting to train-test sets of the cross-validation procedure. We use the 5 models trained per train/test splits instead of retraining the model on the entire data. In sub-challenge 2, we also determine our choice by the variable transformations used. The gain from ensembling is around 0.01 in sub-challenge 2 and 0.04 in sub-challenge 3, as determined on a holdout set.
NCI-CPTAC DREAM-Proteogenomics Challenge – Conclusions
The Outlook
As our solution scored with the second place in sub-challenge 3, we were invited to the community phase, in which we worked on a joint improved solution with the winning team: Hongyang Li and Yuanfang Guan from the University of Michigan. As a result, the performance of the models was significantly improved (from the mean Pearson correlation of 0.33 to 0.37 for the ovarian cancer, and from 0.42 to 0.48 for the breast cancer). At the moment the organizers of the challenge and the top teams of sub-challenges 2 and 3 are finalizing a manuscript on the results of the sub-challenges, which is to be submitted to Nature Methods. More details on the collaboration can be found in [10].Works Cited:
[3] dreamchallenges.org/publications
[4] NCI-CPTAC DREAM Proteogenomics Challenge, www.synapse.org/#!Synapse:syn8228304/wiki/413428
[5] Clinical Proteomic Tumor Analysis Consortium (CPTAC), proteomics.cancer.gov/programs/cptac
[6] Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, et al., Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer, Cell 2016;166: 755–765. doi.org/doi:10.1016/j.cell.2016.05.069
[7] Hastie T, Tibshirani T, Tibshirani RJ, Extended Comparisons of Best Subset Selection, Forward Stepwise Selection, and the Lasso, https://arxiv.org/abs/1707.08692
[8] MSigDB Collections, software.broadinstitute.org/gsea/msigdb/collections.jsp
[9] A resource for Human, Mouse and Rat Transcription Factors, www.tfcheckpoint.org/index.php/introduction
[10] Best Performers Announced for the NCI-CPTAC DREAM Proteogenomics Computational Challenge, proteomics.cancer.gov/news_and_announcements/best-performers-announced-nci-cptac-dream-proteogenomics-computational




