This article discusses Optimizing bioinformatics workflows is critical for reproducibility, efficiency, and agility—especially as datasets and complexity grow., with a case study detailing Genomics England’s successful transition to Nextflow-based pipelines to process 300,000 whole-genome sequencing samples. If you’re attending the Festival of Genomics and Biodata on January 29-30th, we invite you to check out our talk, where we will be presenting similar successful case studies.
Bioinformatics workflows play a pivotal role in managing and analyzing complex biological data. A bioinformatics workflow is a structured series of computational steps designed to process, analyze and interpret data generated from experiments such as next-generation sequencing (NGS). These workflows are essential for transforming raw data into meaningful insights, enabling researchers to identify patterns, make predictions and derive conclusions about biological systems. Whether it’s a small-scale analysis of a single dataset or a large-scale solution for processing terabytes of data, bioinformatics workflows provide the foundation for efficient, reproducible and scalable data management in this increasingly data-driven domain.
When it comes to optimizing bioinformatics workflows, everyone’s needs are different. Some companies might be looking for an optimal solution to evaluate the results of a specific study, such as a large cohort analysis, while others might focus on scalable solutions that simplify daily operations. In either case, setting up bioinformatics workflows in the right way is key to organizational efficiency, improved resource management and faster time-to-insights.
Considering the cost of bioinformatics workflows
The costs associated with bioinformatics workflows can escalate quickly, depending on the scale of analysis. For example, while analyzing a single sample may cost less than $1, processing millions of data points can result in monthly data processing expenses that reach tens or even hundreds of thousands of dollars. Additionally, some companies still rely on semi-manual workflows where full-time employees (FTEs) are tasked with moving data through unstable pipelines, further compounding inefficiencies and increasing costs.
Investing in the optimization of bioinformatics workflows can therefore save companies a lot of capital in the long run, as well as significantly accelerate their operations. An important thing to consider is balancing the cost of optimization and usage. A good rule of thumb is that optimization may take at least two months to complete, but the payoff is substantial, with potential time and cost savings ranging from 30% to 75%.
When should you optimize your bioinformatics workflows?
As your organization grows, bioinformatics processing demands will inevitably scale. Investing in scalable infrastructure early on allows workflows to expand alongside your needs. Optimization should begin when usage scales justify the cost. This process can be divided into three stages:
Analysis Tools: The first and often most challenging step involves identifying and implementing improved analysis tools. This requires exploratory analysis to find tools that fit your specific needs or developing custom solutions if none exist. Focus on addressing the most demanding, unstable or inefficient points first, as these will have the greatest impact.
Workflow Orchestrator: Introducing a dynamic resource allocation system helps prioritize operations based on dataset size. This prevents over-provisioning, reduces computational costs and enhances overall efficiency.
Execution Environment: Ensuring a cost-optimized execution environment is essential, especially for cloud-based workflows. Misconfigurations in cloud setups are common and can lead to unnecessary expenses. Proper environment configuration minimizes these risks and ensures smoother operations.
By approaching optimization in these three stages, organizations can ensure that their bioinformatics workflows remain adaptable and efficient as demands grow. It’s important to remember that optimization is not a one-time process but an ongoing effort to refine tools, orchestrators and environments as data requirements evolve. Starting the optimization process at the right time not only prevents bottlenecks but also sets a strong foundation for scalability, enabling teams to focus on delivering actionable insights rather than managing technical limitations.
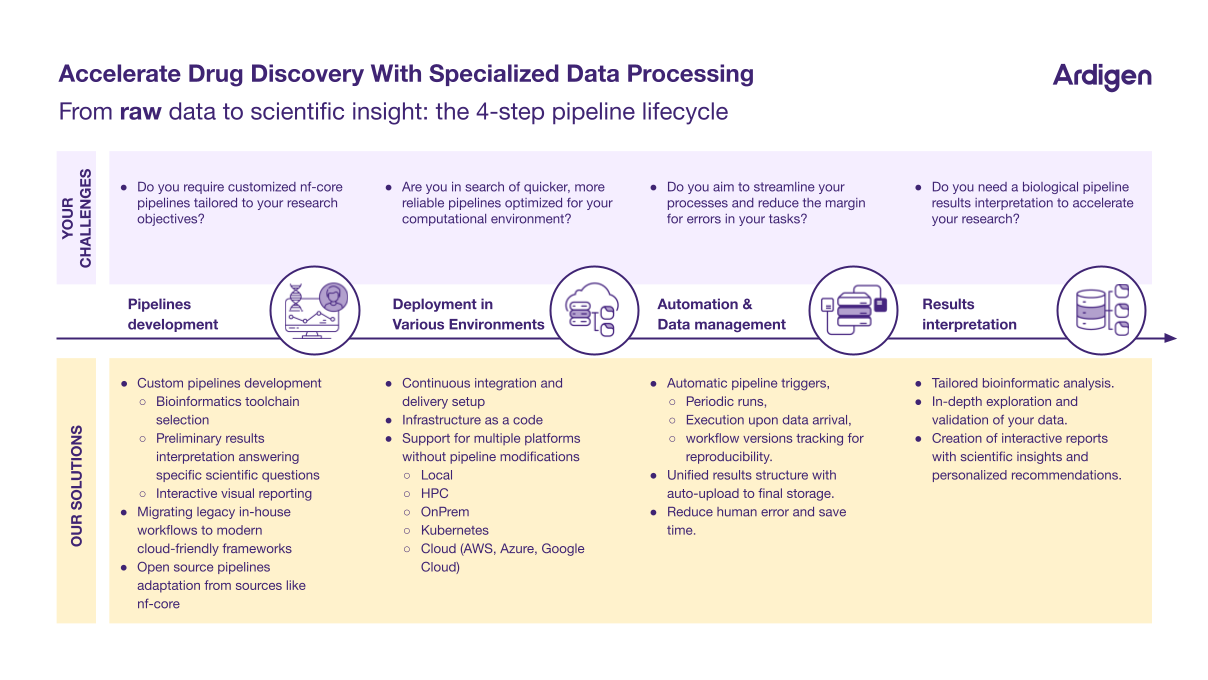
From raw data to scientific insight: the 4-step pipeline lifecycle
Before transitioning your bioinformatics workflows, it is important to assess your needs and goals. Here are four key areas where bioinformatics workflows can be improved:
Pipeline Development: Custom pipeline development and bioinformatics toolchain selection are vital for tailoring workflows to specific research objectives. This includes interpreting preliminary results, generating interactive visual reports, and migrating legacy workflows to modern, cloud-friendly frameworks. Open-source pipelines, such as those from nf-core, can also be adapted to streamline operations.
Deployment Across Environments: Optimized workflows should be deployable across multiple environments, including local machines, high-performance computing (HPC) clusters, on-premise systems, Kubernetes, and cloud platforms (AWS, Azure, Google Cloud). Ensuring compatibility across these platforms without requiring pipeline modifications can improve efficiency and reduce errors.
Automation and Data Management: Automation can significantly streamline workflows. Features such as automatic pipeline triggers, periodic runs, execution upon data arrival, and version tracking for reproducibility reduce human error and save time. Unified result structures with automatic uploads to final storage further enhance efficiency.
Results Interpretation: Beyond data processing, tailored bioinformatics analysis enables in-depth exploration and validation of results. Interactive reports provide actionable scientific insights and personalized recommendations, accelerating research progress.
Case study: Migrating clinical workflows to Nextflow at a national scale
One example of successful workflow optimization is Genomics England’s transition to Nextflow-based pipelines. This project aimed to process 300,000 whole-genome sequencing samples by 2025 for the UK’s Genomic Medicine Service. The migration involved replacing their internal workflow engine with Genie, a solution leveraging Nextflow and the Seqera Platform. Agile methodologies, bi-weekly sprints, and continuous stakeholder feedback ensured a smooth transition while maintaining high-quality outputs through rigorous testing frameworks. The project prioritized patient benefits and scalability within a conservative operational framework. For those attending the Festival of Genomics and Biodata on January 29- 30th, we invite you to check out Ardigen’s talk, where we will be presenting similar successful case studies.
Ensuring a smooth and timely transition
The responsibility for ensuring timely optimization of bioinformatics workflows largely falls on project budget managers, as they oversee resource allocation and long-term cost efficiency. Without proper optimization, workflows can become bottlenecks, consuming excessive time, computational power, and financial resources, especially as data processing demands scale.
By prioritizing optimization at the right stages, budget managers can enable teams to handle larger datasets within the same financial constraints, maximizing the value of available resources. Strategic improvements to workflows can lead to significant cost savings and performance enhancements. This proactive approach allows companies to process larger amounts of data faster on the same budget, accelerating research and discovery progress.
If you want to learn more about how you can optimize your bioinformatics workflows, reach out to one of Ardigen’s specialists who can help you identify areas of improvement and guide you toward more efficient solutions.




