
These organisms – bacteria, viruses, fungi and protozoa – living inside and on the surface of the human body are collectively called microbiota and their genetic material is called microbiome. Most of them are found in the intestines, which make fecal samples the main source of knowledge about the microbiome. Currently, the accuracy and the number of microbiome studies have increased significantly due to the availability of next-generation sequencing. However, before you sequence the microbiome data, you must go through the stool sample processing [8,9].
The process includes collecting, transporting and storing samples, as well as DNA isolation. All of these steps affect the quality of the sample. However, using multiple sample processing methods makes it difficult to compare results between experiments. Therefore, it is very important to standardize the process. Developing detailed Standard Operating Procedures (SOPs) before collecting samples guarantees high quality and reproducibility of the obtained results. A flow chart of an example sample processing is shown below [8,10,12].
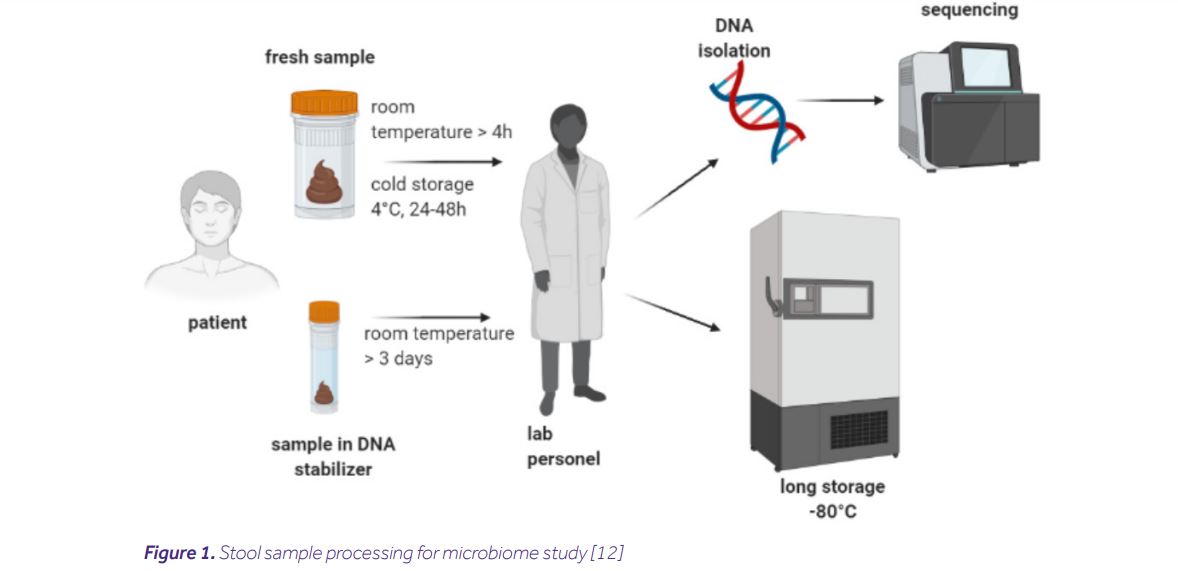
Collecting samples
Collecting stool samples is the first and one of the most important steps in any study of the microbiome. Unfortunately, this procedure is also subject to the greatest number of mistakes. In order to ensure the comfort of patients, collection is most often carried out at their homes by themselves [8]. Due to the fact that it is not an experienced researcher who collects a sample, every effort should be made to simplify this process for the patient. To unify conditions for collecting material, it is necessary to use special kits designed for this purpose. The greatest risk at this stage is the possibility of DNA degradation. Therefore, it is recommended to use sets supplemented with DNA stabilizers [4]. Due to the possibility of the patient making mistakes that can significantly affect the quality of the samples, it is important to inform them about the appropriate amount of collected material and the conditions in which it must be stored before it reaches the laboratory.
The appropriate quality of the sample in the absence of a stabilizer can be achieved through immediate delivery of the material to the laboratory (up to 4 hours), or its storage by the patient in refrigerated conditions until transport. Each sample should be pseudonymised and clearly signed preventing sample confusion and allowing for patient re-identification at the end of the process [6,7,12].
Transport
Transporting conditions are crucial to the procedure. It is essential to transport samples at the same temperature throughout the process to avoid damage and degradation of the samples, due to repeated freezing and thawing. Moreover, the use of an anaerobic atmosphere allows for the additional protection of microorganisms that do not tolerate oxygen. Anaerobic atmosphere generation bags can be skipped when using DNA stabilizers in the previous step. If precautions necessary to maintain the cold chain at every stage of the delivery are not taken, samples will no longer be of value. Additionally, paying attention to protecting against mechanical damage is an important aspect. More often than not, the material is placed in plastic containers that may crack, causing sample loss and contamination of other samples if not handled with precaution. When transported by a patient, it is necessary to secure the sample in a polystyrene container with cooling inserts to prevent temperature increases. However, if the sample is to be transported over long distances, it is necessary to provide professional shipment with controlled cooling
conditions throughout the delivery process [5,8,11,12].
Storage
If long-term storage is required, the sample must unconditionally be placed in the freezer at -80 Celsius. It is crucial that the storage conditions are controlled on an ongoing basis and the temperature is kept constant. Temperature fluctuations have a deteriorating effect on the quality of the samples. Although storing the sample seems easy enough, there are things that can go wrong also during this stage. Potential problems may occur as a result of equipment’s power supply failure, which can cause thawing and thus loss of all samples. Mechanical damage may also occur. This is often due to improper storage, overloading the freezer or containers made of poor quality materials. To avoid damaging the containers and the contamination of samples, it needs to be assured that the container used to collect the sample is suitable for storage in low temperatures. Apart from that, it is crucial to label the containers in such a way that the label does not blur due to freezing and thawing. It is recommended to use special labels adapted to this process, instead of labeling the containers with a marker [1,2,4,5,7].
DNA isolation
If all the previous stages of sample processing have been carried out without complications, the DNA isolation may proceed. Note that when using a frozen sample, it must be thawed first. The stool samples are not homogeneous, therefore thorough homogenization is required every time. In its absence, the portion used for isolation may not be representative of the whole sample causing incorrect results down-the-line. The key issue in performing the DNA isolation is the selection of an appropriate kit that meets our requirements for the desired output. To achieve reproducible results, it is necessary to proceed step by step in accordance with a previously prepared Standard Operating Procedure. Good laboratory practice must also be strictly adhered to. Even the slightest neglect can cause the contamination of samples with researcher’s DNA. The experience of the researcher as well as accuracy and careful performance of subsequent activities are highly important at this stage. The process must be carried out efficiently, so that the tested sample does not deteriorate. Leaving the samples in inappropriate conditions has a very negative impact on the process and the quality of the obtained DNA. Until isolation begins, samples should be kept on dry ice and after thawing, immediately collected, analyzed and the rest of the material closed and secured. To avoid failures, after the isolation process, the quality of the obtained DNA and quantity in the sample should be checked. Following all the procedures and taking all precautions will allow you to obtain DNA of a good quality, which is the basis for performing next-generation sequencing and obtaining valuable information. This will allow further bioinformatic processing [3,6,8,10,12].
You could probably see by now that if the procedures are not followed in the subsequent stages of sample processing, obtained data would be of poor quality or the DNA sequencing will not be possible at all. Standardized sample processing ensures better data quality and allows to obtain more information contained in the test material. Therefore, before starting the study, it is worth considering and planning the process and preparing the necessary documentation well in advance to improve its flow and guarantee success.
Our goal at Ardigen is to control each stage of the process as the proper data generation is a crucial step for data-driven projects in the field of healthcare. We have seen multiple projects where even simple changes, such as choosing a different isolation kit or slight adjustments to the sequencing protocol, resulted in major improvement in the quality of produced results. This experience is an important value we bring into each project.
At Ardigen, we pay much attention to the details at each step of the microbiome research process, which allows us to work smoothly and effectively. Stay up to date with our latest projects and news by subscribing to our newsletter at microbiome.ardigen.com.
Bibliography
[1] Blekhman R, Tang K, Archie EA, Barreiro LB, Johnson ZP, Wilson ME, et al. Common methods for fecal sample storage in field studies yield consistent signatures of individual identity in microbiome sequencing data. Sci Rep. 201;6: 31519.
[2] Choo J, Leong L, Rogers G. Sample storage conditions significantly influence faecal microbiome profiles. Sci Rep. 2015;5; 16350.
[3] Costea PI, Zeller G, Sunagawa S, et al., Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol, 2017. 35(11): 1069-1076.
[4] Flores R, Shi J, Yu G, Ma B, Ravel J, Goedert JJ, Sinha R. Collection media and delayed freezing effects on microbial composition of human stool. Microbiome. 2015;3:33.
[5] Fouhy F, Deane J, Rea MC, O’Sullivan Ó, Ross RP, O’Callaghan G, et al. The Effects of Freezing on Faecal Microbiota as Determined Using MiSeq Sequencing and Culture-Based Investigations. PLOS ONE. 2015;10:e0119355.
[6] Gorzelak MA, Gill SK, Tasnim N, Ahmadi-Vand Z, Jay M, Gibson DL. Method for improving Human Gut Microbiome Data by Reducing Variability through Sample Processing and Storage of Stool. PLOS ONE. 2015;10(9); e0139529 (homogenizacja)
[7] Kait F. Al, Jordan E. Bisanz, Gregory B. Gloor, Gregor Reid, Jeremy P. Burton, Evaluation of sampling and storage procedures on preserving the community structure of stool microbiota: A simple at-home toilet-paper collection method, 2018; 144;117-121
[8] Kumar, R, Eipers E, Little R, Crowley M, et al., Getting started with microbiome analysis: sample acquisition to bioinformatics. Current protocols in human genetics, 2014; 82: p. 18.8.1-18.8.29.
[9] Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacteria to Host Cells in Humans. Cell. 2016; 164;337-40
[10] Sinha, R, Chen J, Amir A, Vogtmann E, et al., Collecting fecal samples for microbiome analyses in epidemiology studies. Cancer Epidemiology and Prevention Biomarkers, 2016; 25(2): 407-416
[11] Song SJ, Amir A, Metcalf JL, Amato KR, Xu ZZ, Humphrey G, Knight R. Preservation Methods Differ in Fecal Microbiome Stability, Affecting Suitable for Field Studies. mSystem. 2016;1.
[12] Wu W.-K., Chen C.-C., Panyod S., Chen R.-A., Wu M.-S., Sheen L.-Y., Chang S.-C. Optimization of fecal sample processing for microbiome study — The journey from bathroom to bench. Journal of the Formosan Medical Association. 2019; 118(2). 545-555




