
While Robinson’s dictum holds for all diseases, it rings particularly true for cancer patients. Cancer is as diverse as the patients who bear it. Even if two patients have the same type of cancer, their particular mutations may differ, and different therapeutic approaches may be required. There is also another issue at stake—cancer is a moving target. As they grow, tumours pick up new mutations that might affect its metabolism and response to treatment. This means that a treatment that is effective today might not be so in just a few weeks. In the end, the therapeutic approach ought to be reassessed regularly.
Advances in biotechnology have brought more targeted and personalized treatments to the clinic. While there are no truly identical cancers, most share the same dangerous trait—they can trick the host’s immune system into ignoring them. In recent years, we have learned how to lift the veil and make tumors vulnerable to our own defences. Immunotherapy drugs such as atezolizumab unblock immune cells and allow them to attack the tumor.
Immunotherapy holds great promise, especially for several difficult-to-treat cancers, such as acute and chronic lymphocytic leukemia. [1] However, there are trade-offs to consider. There are risks involved in mobilizing the immune system to fight cancerous formations. Immunotherapy has been reported to cause serious side effects ranging [2] from fatigue and decreased appetite to severe infections. Additionally, the price of these treatments is significant. For example, in the US, the standard 7.5-month-long [3] treatment with atezolizumab costs almost $100,000 and is not fully covered by insurers. Combined with an average two-year patient survival of 28% [4], this has limited the adoption of these therapies.
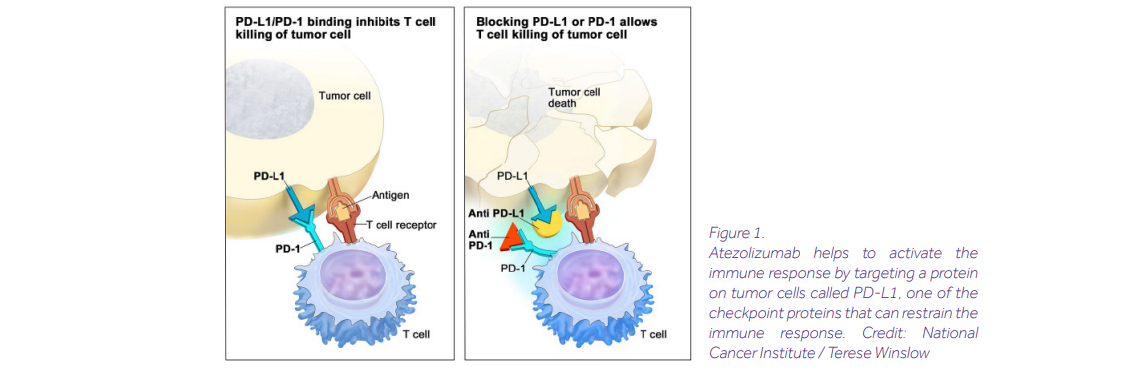
Immunotherapy can only be viable and cost-effective if we know more about the patient’s body and their cancer before making treatment decisions. Determining which patients will benefit from immunotherapy treatments will be critical for the adoption and further development of these therapies.
Identifying biological indicators– biomarkers–that can predict treatment response accurately is almost within our grasp. Vital information lies hidden beneath heaps of data from genomic and transcriptomic panels, immunological assays or in the gut microbiome. Physicians can utilize this knowledge to predict the effectiveness of different therapies in individuals or groups, as well as the likelihood of adverse reactions. Currently, parameters such as the tumour mutational burden (TMB) [5], and Programmed cell death-ligand 1 Immunochemistry (PD-L1 IHC) [6] can serve as predictors of treatment outcome.
Still, considering more factors influencing response is needed to make more accurate predictions. However, the amount of data required to extract relevant insights is too large to be parsed on the fly by healthcare professionals. Certainly, tracking and comparing the data on many different biomarkers to infer a recommendation for therapeutic approach, requires more advanced computational methods and models. These models should also enable the clinicians to interpret the model predictions to justify treatment recommendations.
Biomarker discovery is a growing field and new tools for outcome prediction and diagnostics are rapidly emerging. At Ardigen, we are the forefront of this exploration. We have applied our Bioinformatics and Machine Learning expertise to sift through different sources of data– from genomics, through metabolomics, to biopsy images – and provide insights for a wide variety of diseases ranging from depression to inflammatory bowel disease. To date, we have completed ten biomarker discovery projects.
In oncology, we have built models to understand the contributions of multiple biomarkers for predicting response to atezolizumab. [7] We investigated associations of gene mutations, expressions, and gene expression signatures with the treatment outcome, which lead to the identification of the most relevant biomarkers. We constructed multivariate machine learning models for predicting treatment response. These models were tested on two tasks related to medical outcome measures: one related to tumour size change, and the second one related to patient’s survival during two years post-treatment. In the latter we performed stratification of patients into groups predicted to benefit and not benefit from the atezolizumab treatment.

Compared with standard approaches based on a single biomarker, Ardigen’s model can distinguish which patients will benefit from treatment more sharply. Our model can discern between two groups of patients: those who after two years will have a survival probability surpassing 60% and those with a likelihood of survival of less than 25%. For the latter group, early information about the low effectiveness of the potential treatment can mean that other options might be pursued to preserve health and life. Thus, the model can guide physicians in matching up patients with the most beneficial treatments.
Every single one of the 7.59 billion human genomes is unique, even those of identical twins [8]. We all have genes that make us vulnerable to specific diseases and respond differently to treatments. The therapeutic approaches developed in the past century were never designed to take this diversity of genomes into account, limiting the efficacy of treatment. The one-size-fits-all approach that has carried us so far is now giving diminishing returns. Good physicians –like Prof. Andrew Robinson– always placed the individual characteristics of their patients front and center. The remarkable development of methods that reveal the uniqueness of patients at the deepest levels presents these physicians with a new opportunity. Matching the right patient to the right care might soon be a reality.
Bibliography
[1] Brentjens, Renier J. at al. “CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia” (2013) https://pubmed.ncbi.nlm.nih.gov/23515080/
[2] https://www.tecentriq.com/
[3] Morris, Emma, “Roche’s Tecentriq ®▼ (atezolizumab) not approved for triple negative breast cancer” https://pharmafield.co.uk/pharma_news/tecentriq-by-roche-not-approved-on-nhs-for-triple-negative-breast-cancer/#:~:text=The%20cost%20of%20a%20course,the%20NHS%20a%20confidential%20discount.
[4] von Pawel, J. at al. “Long-term survival in patients with advanced non–small-cell lung cancer treated with atezolizumab versus docetaxel: Results from the randomised phase III OAK study” (2019) https://www.sciencedirect.com/science/article/abs/pii/S0959804918315181#:~:text=Patients%20with%20the%20highest%20level,and%2017.4%25%2C%20respectively%20
[5] Boumber, Yanis, “Tumor mutational burden (TMB) as a biomarker of response to immunotherapy in small cell lung cancer” (2018) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6129910/
[6] Scheel, Andreas H. and Schäfer, Stephan C. “Current PD-L1 immunohistochemistry for non-small cell lung cancer” (2018) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5906363/
[7] Shah, Parantu K. at al. “Understanding contribution and independence of multiple biomarkers for predicting response to atezolizumab” (2019) https://ascopubs.org/doi/abs/10.1200/JCO.2019.37.15_suppl.2567
[8] Casselman, Anne, “Identical Twins’ Genes Are Not Identical” (2008) https://www.scientificamerican.com/article/identical-twins-genes-are-not-identical/
Further Reading from Ardigen’s Knowledge Hub
Dive deeper into how AI and advanced bioinformatics are transforming precision medicine and drug discovery:




