What if you could uncover how to treat the disease without knowing the target? Integrating phenotypic screening with omics and AI offers just that.
For decades, target-based drug discovery has dominated the pharmaceutical landscape. However, biology does not always follow linear rules. The resurgence of phenotypic screening signals a shift back to a biology-first approach, made exponentially more powerful by modern omics data and AI.
Why Phenotypic Screening Is Changing the Game
Phenotypic screening allows researchers to observe how cells or organisms respond to genetic or chemical perturbations without presupposing a target. With advancements in high-content imaging, single-cell technologies, and functional genomics (e.g., Perturb-seq), this approach now captures subtle, disease-relevant phenotypes at scale. The result? Unbiased insights into complex biology.
Three trends make this possible:
- Data richness: multiplexed assays, single-cell sequencing, and automated imaging offer multi-dimensional phenotypic profiles (Soule et al., 2024; Bonnar et al., 2021; Singh et al., 2022; Loh et al., 2017; Hahn et al., 2023).
- Scalability: new methods pool perturbations and use computational deconvolution, dramatically reducing sample size, labor, and cost while maintaining information-rich outputs (Soule et al., 2024; Hahn et al., 2023).
- Computational power: AI and machine learning models interpret massive, noisy datasets to detect meaningful patterns (Loh et al., 2017; Brown & Wobst, 2020).
Omics Layers Add Biological Context
Genomics, transcriptomics, proteomics, metabolomics, and epigenomics reveal a piece of the phenotype puzzle. Multi-omics approaches focus on integrating them. Researchers gain a systems-level view of biological mechanisms that single-omics analyses cannot detect.
- Transcriptomics reveals active gene expression patterns.
- Proteomics clarifies signaling and post-translational modifications.
- Metabolomics contextualizes stress response and disease mechanisms.
- Epigenomics gives insights into regulatory modifications.
Multi-omics integration improves prediction accuracy, target selection, and disease subtyping, which is critical for precision medicine (Binder et al., 2022; Sharma et al., 2024; Zhou et al., 2024).
The Growing Role of AI in Data Integration
AI/ML models enable the fusion of multimodal datasets that were previously too complex to analyze together. Deep learning and interpretable models can:
- Combine heterogeneous data sources (e.g., electronic health records, imaging, multi-omics, sensor data) into unified models (Moorthy et al., 2025; Ali et al., 2022; Koch et al., 2021; Mgbole, 2025; Moshawrab et al., 2023).
- Enhance predictive performance in disease diagnosis, particularly early cancer detection, and biomarker discovery (Ali et al., 2022; Raghunathan et al., 2024; Mgbole, 2025; Alwafai et al., 2022).
- Personalize therapies with adaptive learning from patient data (Moorthy et al., 2025; Mendhe et al., 2024).
Applications range from cancer biomarker discovery to drug response prediction in CNS disorders. Tools like IntelliGenes and ExPDrug exemplify how AI platforms make integrative discovery accessible to non-experts.
Drug Candidates Uncovered Through Integration
You do not always need to know the destination if you have the right map. Several promising candidates in oncology and immunology were identified not through target-based screening but by computational backtracking of observed phenotypic shifts.
Notable examples include:
- Lung cancer: Archetype™ AI identified AMG900 and new invasion inhibitors using patient-derived phenotypic data, along with omics (Neyarapally et al., 2025).
- COVID-19: The DeepCE model predicted gene expression changes induced by novel chemicals, enabling high-throughput phenotypic screening for COVID-19. This approach generated new lead compounds consistent with clinical evidence, demonstrating the power of integrating phenotypic and omics data with AI for rapid drug repurposing (Pham et al., 2021).
- Triple-negative breast cancer: the idTRAX machine learning-based approach has been used to identify cancer-selective targets (Gautam et al., 2018).
- Antibacterial discovery: GNEprop and PhenoMS-ML models uncovered novel antibiotics by interpreting imaging and mass spec phenotypes (Scalia et al., 2024; Oosten & Klein, 2020).
These examples demonstrate how integrative platforms reduce timelines and enhance confidence in hit validation.
Challenges: The Data Is not Always Ready
While the science is compelling, practical barriers remain:
- Data heterogeneity and sparsity: different formats, ontologies, and resolutions complicate integration. Additionally, many datasets are incomplete or too sparse for the effective training of advanced AI models, particularly in fields such as oncology (Khosravi et al., 2021).
- Privacy & ethics: sensitive health data requires compliance and debiasing.
- Interpretability: deep learning and complex AI models often lack transparency, making it difficult for clinicians to interpret predictions and trust the results.
- Infrastructure: multi-modal AI demands large datasets and high computing resources, so it remains a technical hurdle.
Efforts like FAIR data standards, open biobank initiatives, and user-friendly ML toolkits are addressing these gaps.
Looking Ahead
Integrating phenotypic data with omics and AI is not just an upgrade. It is a new operating system for drug discovery. By starting with biology, adding molecular depth, and letting algorithms reveal the patterns, we are moving towardeffective and better-understood therapies.
At Ardigen, we have built PhenAID to bridge the gap between advanced phenotypic screening and actionable insights. This AI-powered platform integrates cell morphology data, omics layers, and contextual metadata to identify phenotypic patterns that correlate with the mechanism of action, efficacy, or safety.
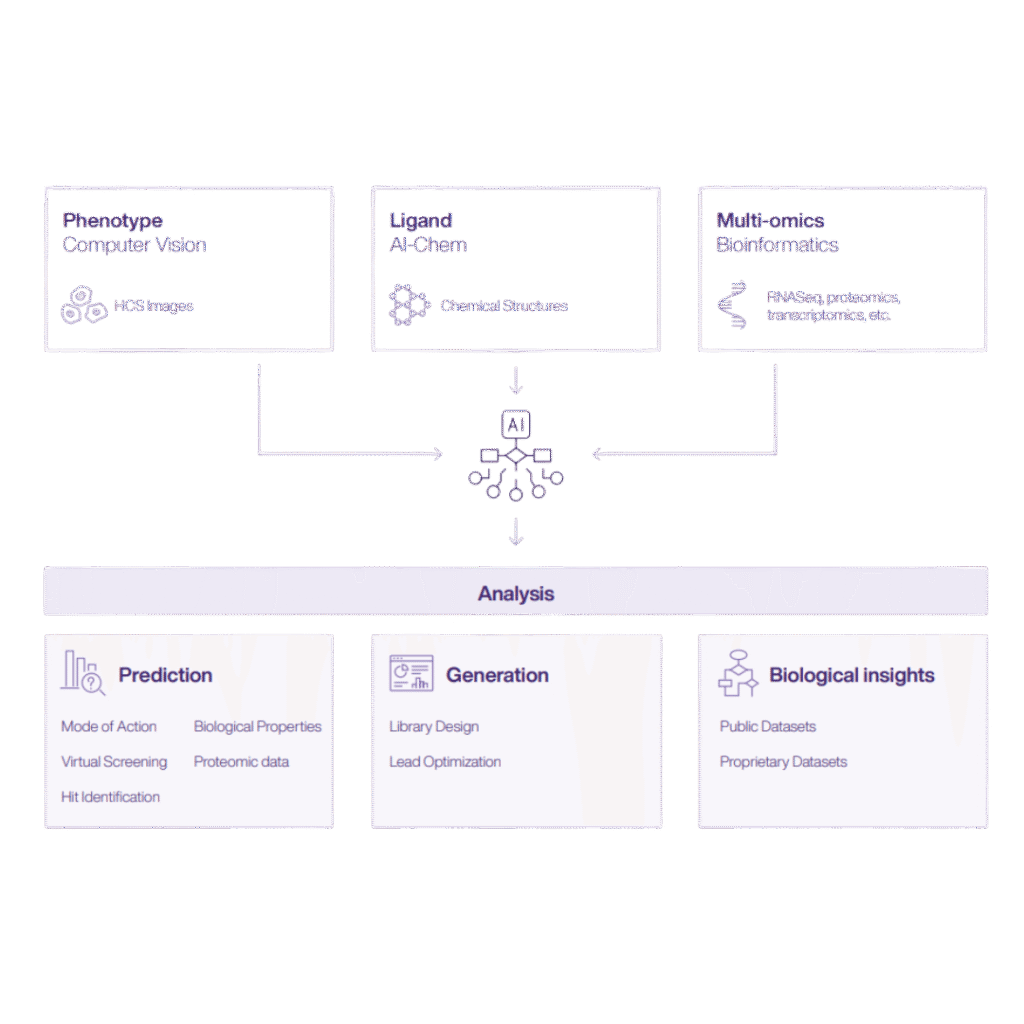
The core of the application utilizes high-content data from microscopic images obtained with the Cell Painting assay, which visualizes important cellular components or organelles (Bray et al., 2016). Image analysis pipelines paired with robust data preprocessing enable the detection of subtle changes in cell morphology and generate profiles that are further compared to identify biologically active compounds.
Additional capabilities support further characterization:
- The bioactivity prediction module integrates multimodal data to characterize compounds or predict their on and off-target activity.
- The MoA (Mechanism of Action) prediction tool elucidates how tested compounds may interact in the biological setting.
- Virtual Screening identifies compounds that induce a desired phenotype, thereby accelerating viable drug candidate identification and reducing lab costs.
By combining AI and human expertise, PhenAID empowers researchers to decode phenotypic complexity and fast-track the road from image to insight. The platform is already being used in collaborations to uncover new drug targets and refine lead compounds across oncology, immunology, and infectious diseases.
The future of drug discovery is already underway.
Author: Martyna Piotrowska |
Technical editing: Ardigen expert Magdalena Otrocka
Bibliography:
Soule, C., Cleary, B., Kummerlowe, C., Mead, B., Cheah, J., Vrcic, A., Lowder, K., Shalek, A., Bryson, B., Triana, S., Crawford, L., Dao, T., Raghavan, S., Amini, A., Blainey, P., Guzman, M., Winter, P., Hahn, W., Peters, J., Cheng, T., McIninch, J., Kattan, W., Liu, N., & Ingabire, S. (2024). Scalable, compressed phenotypic screening using pooled perturbations. Nature biotechnology. https://doi.org/10.1038/s41587-024-02403-z
Bonnar, J., Iremadze, N., Lithwick-Yanai, G., Hussmann, J., Pogson, A., Jost, M., Adelman, K., Mascibroda, L., Replogle, J., Saunders, R., Weissman, J., Guna, A., Oberstrass, F., LeNail, A., Lipson, D., Norman, T., & Wagner, E. (2021). Mapping information-rich genotype-phenotype landscapes with genome-scale Perturb-seq. Cell, 185, 2559 – 2575.e28. https://doi.org/10.1016/j.cell.2022.05.013
Singh, A., Funk, L., Blainey, P., Soong, B., Leiken, M., Carlson, R., Feldman, D., Le, A., & Tsai, F. (2022). Pooled genetic perturbation screens with image-based phenotypes. Nature Protocols, 17, 476–512. https://doi.org/10.1038/s41596-021-00653-8
Loh, S., Bougen-Zhukov, N., Loo, L., & Lee, H. (2017). Large‐scale image‐based screening and profiling of cellular phenotypes. Cytometry Part A, 91. https://doi.org/10.1002/cyto.a.22909
Hahn, W., Raghavan, S., Soule, C., Winter, P., Lowder, K., Bryson, B., Cleary, B., Kummerlowe, C., Mead, B., Peters, J., Cheng, T., Kattan, W., Cheah, J., Blainey, P., Liu, N., & Shalek, A. (2023). Compressed phenotypic screens for complex multicellular models and high-content assays. bioRxiv. https://doi.org/10.1101/2023.01.23.525189
Wagner, B., & Schreiber, S. (2016). The Power of Sophisticated Phenotypic Screening and Modern Mechanism-of-Action Methods. Cell chemical biology, 23 1, 3-9. https://doi.org/10.1016/j.chembiol.2015.11.008
Brown, D., & Wobst, H. (2020). Opportunities and challenges in phenotypic screening for neurodegenerative disease research. Journal of medicinal chemistry. https://doi.org/10.1021/acs.jmedchem.9b00797
Binder, H., Schmidt, M., Hopp, L., Arakelyan, A., Davitavyan, S., & Loeffler-Wirth, H. (2022). Integrated Multi-Omics Maps of Lower-Grade Gliomas. Cancers, 14. https://doi.org/10.3390/cancers14112797
Sharma, G., Banerjee, S., Chakraborty, S., & Karmakar, S. (2024). Multi-OMICS approaches in cancer biology: New era in cancer therapy. Biochimica et biophysica acta. Molecular basis of disease, 167120. https://doi.org/10.1016/j.bbadis.2024.167120
Zhou, Z., Zhang, Y., Xu, Y., Han, X., Zou, H., Weng, S., Liu, Z., Lin, T., Zhou, A., Zhang, G., & Chen, S. (2024). Omics-based molecular classifications empowering in precision oncology. Cellular oncology. https://doi.org/10.1007/s13402-023-00912-8
Moorthy, A., Leena, G., Sangeetha, S., Gayathri, R., Mathivanan, S., Sangeetha, R., & Mary, R. (2025). Dynamic AI-Enhanced Therapeutic Framework for Precision Medicine Using Multi-Modal Data and Patient-Centric Reinforcement Learning. IEEE Access, 13, 77709–77733. https://doi.org/10.1109/ACCESS.2025.3564971
Ali, H., Hajj, N., Mohsen, F., & Shah, Z. (2022). Artificial intelligence-based methods for fusion of electronic health records and imaging data. Scientific Reports, 12. https://doi.org/10.1038/s41598-022-22514-4
Akhtar, S. (2025). Multi-modal Generative AI Models: Architecture, Benefits, Applications, and Challenges. International Journal of Advanced Research in Science, Communication and Technology. https://doi.org/10.48175/ijarsct-24447
Koch, W., Leung, H., Chong, C., Blasch, E., Braines, D., Abdelzaher, T., & Pham, T. (2021). Machine Learning/Artificial Intelligence for Sensor Data Fusion–Opportunities and Challenges. IEEE Aerospace and Electronic Systems Magazine, 36, 80–93. https://doi.org/10.1109/MAES.2020.3049030
Raghunathan, T., Mahur, A., B., B., & Mishra, A. (2024). Multi-Modal AI/ML Integration for Precision Glaucoma Detection: A Comprehensive Analysis using Optical Coherence Tomography, Fundus Imaging, RNFL, and Vessel Density. 2024 2nd International Conference on Artificial Intelligence and Machine Learning Applications Theme: Healthcare and Internet of Things (AIMLA), 1–7. https://doi.org/10.1109/AIMLA59606.2024.10531451
Mgbole, T. (2025). Machine learning integration for early-stage cancer detection using multi-modal imaging analysis. World Journal of Advanced Research and Reviews. https://doi.org/10.30574/wjarr.2025.25.1.0066
Mendhe, D., Ahmed, Z., DeGroat, W., Narayanan, R., & Abdelhalim, H. (2024). IntelliGenes: Interactive and user-friendly multimodal AI/ML application for biomarker discovery and predictive medicine. Biology Methods & Protocols, 9. https://doi.org/10.1093/biomethods/bpae040
Moshawrab, M., Ibrahim, H., Adda, M., Bouzouane, A., & Raad, A. (2023). Reviewing Multimodal Machine Learning and Its Use in Cardiovascular Diseases Detection. Electronics. https://doi.org/10.3390/electronics12071558
Peker, E., DeGroat, W., Zeeshan, S., Title, R., Narayanan, R., Mendhe, D., & Ahmed, Z. (2025). 3D IntelliGenes: AI/ML application using multi-omics data for biomarker discovery and disease prediction with multi-dimensional visualization. https://doi.org/10.1101/2025.03.25.25324634
Alwafai, Z., Rauch, G., Gomez, C., Gonçalo, M., Golatta, M., Kapetas, P., Heil, J., Gruber, I., Nees, J., Barr, R., Stieber, A., Wojcinski, S., Balleyguier, C., Schuessler, M., Riedel, F., Duda, V., Pfob, A., Schaefgen, B., Clevert, D., Lu, S., Tozaki, M., Rutten, M., Fastner, S., Ohlinger, R., Sidey-Gibbons, C., Hahn, M., Hennigs, A., Xu, C., & Togawa, R. (2022). The importance of multi-modal imaging and clinical information for humans and AI-based algorithms to classify breast masses (INSPiRED 003): an international, multicenter analysis. European Radiology, 32, 4101 – 4115. https://doi.org/10.1007/s00330-021-08519-z
Li, Y., He, R., Wang, F., Zhou, Y., Cheng, J., Cen, X., Gai, B., Yi, C., Gao, F., Wu, Q., Ding, J., & Liu, J. (2025). Challenges in AI-driven Biomedical Multimodal Data Fusion and Analysis.. Genomics, proteomics & bioinformatics. https://doi.org/10.1093/gpbjnl/qzaf011
Bakare, O. (2025). AI-Driven Multi-Omics Integration for Precision Medicine in Complex Disease Diagnosis and Treatment. International Journal of Research Publication and Reviews. https://doi.org/10.55248/gengpi.6.0125.0650
Martínez-García, M., & Hernández-Lemus, E. (2022). Data Integration Challenges for Machine Learning in Precision Medicine. Frontiers in Medicine, 8. https://doi.org/10.3389/fmed.2021.784455
Moorthy, A., Leena, G., Sangeetha, S., Gayathri, R., Mathivanan, S., Sangeetha, R., & Mary, R. (2025). Dynamic AI-Enhanced Therapeutic Framework for Precision Medicine Using Multi-Modal Data and Patient-Centric Reinforcement Learning. IEEE Access, 13, 77709–77733. https://doi.org/10.1109/ACCESS.2025.3564971
Ahmed, Z. (2020). Practicing precision medicine with intelligently integrative clinical and multi-omics data analysis. Human Genomics, 14. https://doi.org/10.1186/s40246-020-00287-z
Khosravi, P., Boehm, K., Shah, S., Gao, J., & Vanguri, R. (2021). Harnessing multimodal data integration to advance precision oncology. Nature Reviews Cancer, 22, 114–126. https://doi.org/10.1038/s41568-021-00408-3
Chen, R., Mahmood, F., & Lu, M. (2025). Abstract 2460: Judith: An agentic AI system for biomedical image analysis and scientific discovery in precision oncology. Cancer Research. https://doi.org/10.1158/1538-7445.am2025-2460
Shi, H., Zuo, F., Liu, X., He, X., & Jing, J. (2022). Artificial intelligence-based multi-omics analysis fuels cancer precision medicine. Seminars in cancer biology. https://doi.org/10.1016/j.semcancer.2022.12.009
Zhong, Y., Shi, W., Gloster, L., Lais, P., Isgut, M., Swain, A., Giuste, F., Sun, J., Tong, L., & Wang, M. (2023). Integrating Multi-Omics Data With EHR for Precision Medicine Using Advanced Artificial Intelligence. IEEE Reviews in Biomedical Engineering, 17, 80–97. https://doi.org/10.1109/RBME.2023.3324264
Mukhopadhyay, A., & Acharya, D. (2024). A comprehensive review of machine learning techniques for multi-omics data integration: challenges and applications in precision oncology. Briefings in functional genomics. https://doi.org/10.1093/bfgp/elae013
Neyarapally, T., Hawley, J., Sinha, A., Faraby, N., Cashorali, T., Powell, C., McDonagh, P., & Oh, W. (2025). Abstract 1139: Massive scale phenotypic screening using generative chemogenomics repurposes safe drugs and discovers new drugs for intercepting early-stage lung adenocarcinoma progression. Cancer Research. https://doi.org/10.1158/1538-7445.am2025-1139
Pham, T., Xie, L., Zhang, P., Qiu, Y., & Zeng, J. (2021). A deep learning framework for high-throughput mechanism-driven phenotype compound screening and its application to COVID-19 drug repurposing. Nature machine intelligence, 3, 247–257. https://doi.org/10.1038/s42256-020-00285-9
Gautam, P., Jaiswal, A., Aittokallio, T., Al-Ali, H., & Wennerberg, K. (2019). Phenotypic Screening Combined with Machine Learning for Efficient Identification of Breast Cancer-Selective Therapeutic Targets. Cell chemical biology, 26(7), 970–979.e4. https://doi.org/10.1016/j.chembiol.2019.03.011
Scalia, G., Rutherford, S. T., Lu, Z., Buchholz, K. R., Skelton, N., Chuang, K., Diamant, N., Hütter, J.-C., Luescher, J.-M., Miu, A., Blaney, J., Gendelev, L., Skippington, E., Zynda, G., Dickson, N., Koziarski, M., Bengio, Y., Regev, A., Tan, M.-W., & Biancalani, T. (2024). Deep learning enables discovery of novel antibacterial compounds from phenotypic screens [Preprint]. bioRxiv. https://doi.org/10.1101/2024.09.11.612340
van Oosten, L. N., & Klein, C. D. (2020). Machine Learning in Mass Spectrometry: A MALDI-TOF MS Approach to Phenotypic Antibacterial Screening. Journal of medicinal chemistry, 63(16), 8849–8856. https://doi.org/10.1021/acs.jmedchem.0c00040
Bray, M. A., Singh, S., Han, H., Davis, C. T., Borgeson, B., Hartland, C., Gustafsdottir, S. M., Gibson, C. C., & Carpenter, A. E. (2016). Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nature Protocols, 11(9), 1757-1774. https://doi.org/10.1038/nprot.2016.105




